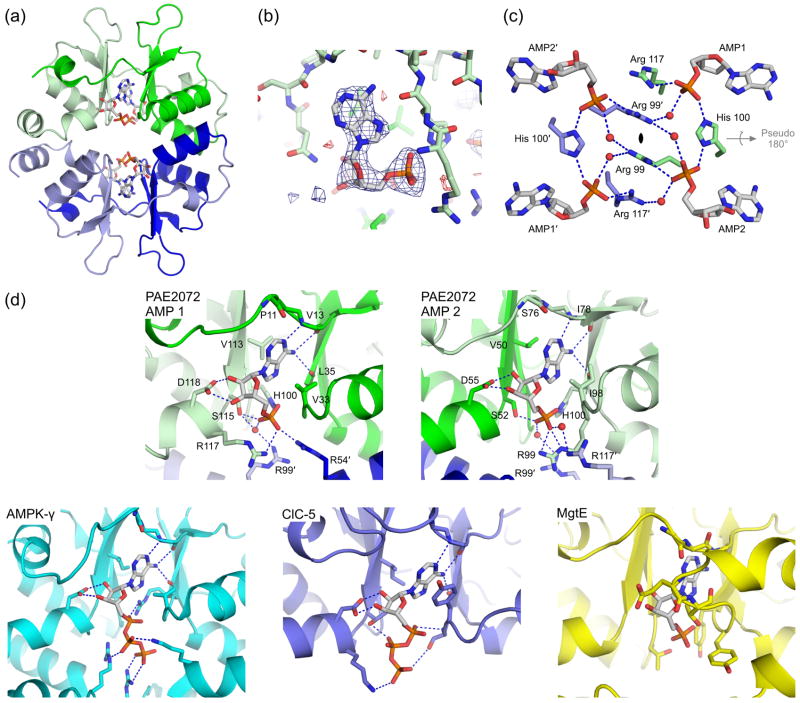
Figure 3. Ligand binding by PAE2072.
(a) A view of one of the two crystallographically independent dimers in the AMP-bound structure shows the positions of the four bound AMP molecules. Note the lack of any significant conformational change from the unliganded structure in Figure 1. (b) Difference density from a simulated annealing Fo-Fc omit map, contoured at 3.0 σ, reveals readily interpretable density for AMP. The coordinates from the final model are shown in stick representation. (c) Three residues from each subunit (Arg99, His100, and Arg117) are involved in coordinating multiple AMP molecules, which may suggest cooperative binding. Several well-ordered water molecules form bridging interactions, and may indicate space where additional phosphate groups, such as those on ATP, could be accommodated. The dimeric two-fold axis is indicated in the center of the figure and runs perpendicular to the page; the pseudo two-fold is shown in gray. (d) The mechanism of adenine nucleotide binding is strongly conserved in the CBS domains of PAE2072 (both binding sites are shown), mammalian AMPK (PDB code 2V92), and human ClC-5 (PDB code 2J9L), but not in bacterial MgtE (PDB code 2YVY). The four central beta strands of one subunit of PAE2072 were aligned with those of the tandem CBS domains of MgtE (RMSD = 0.81 Å over 20 Cα atoms) to model the potential position of a bound AMP molecule. Strong steric clashes between the protein and the modeled AMP can be seen, and no basic residues are in position to coordinate the phosphate residues. It therefore seems unlikely that MgtE binds adenine nucleotides in a manner similar to that observed in the other CBS domains shown here.