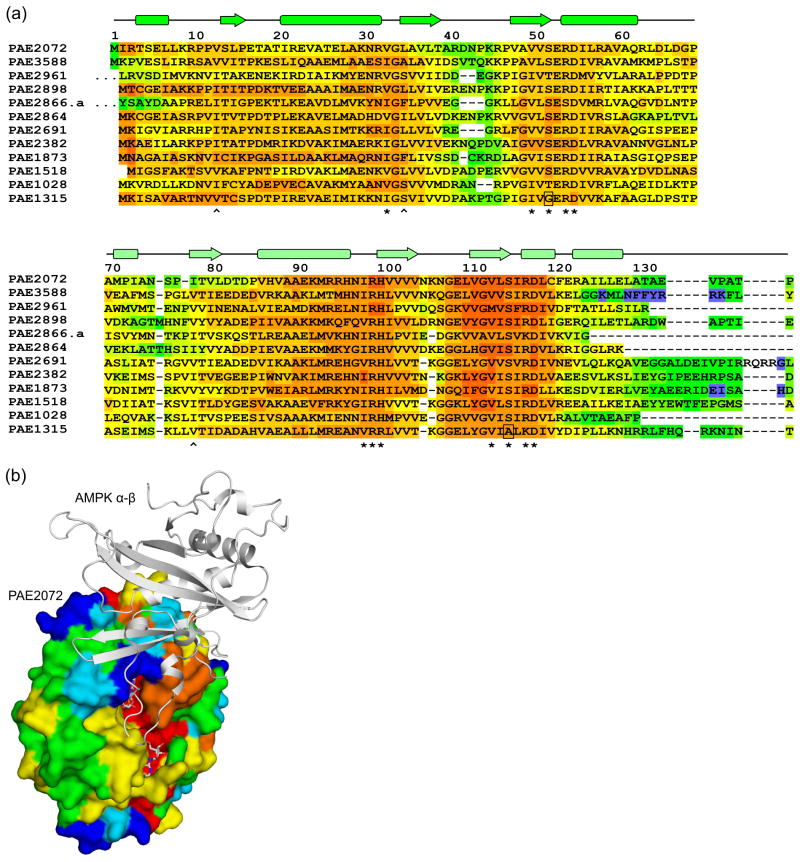
Figure 4. Sequence-structure analysis of Pyrobaculum CBS domain proteins.
(a) A multiple sequence alignment of 12 standalone CBS domain proteins from P. aerophilum, generated using the alignment program T-COFFEE, revealed nearly universal conservation of the residues involved in adenylate binding in PAE2072. Residues with side chains (*) or backbone atoms (^) that contact AMP are indicated. Two substitutions likely to result in the loss of a contact are boxed. The sequences of two regions other than the termini were found to be highly variable in this family of proteins (residues 40–46 and 71–84 in PAE2072). The first 10 residues of PAE2961 and 27 residues of PAE2866.a are represented by ellipses for clarity. (b) The regions of high sequence variability in Pyrobaculum CBS domain proteins map to a single surface patch on PAE2072 which coincides precisely with the site of interaction between the γ subunit of AMPK and the α and β subunits. The four central beta strands of PAE2072 were aligned with those of the γ subunit of S. pombe AMPK (PDB ID 2OOX; RMSD = 0.88 over 20 Cα atoms) to model PAE2072 in the place of the γ subunit. The surface of PAE2072 is shown and colored according to sequence conservation; red indicates highly conserved positions, while blue indicates highly variable positions. The α and β subunits of AMPK are shown in cartoon representation and can be seen to dock onto the surface patch defined by the highly variable segments (blue and cyan). The AMP molecules bound to PAE2072 are shown in sticks to emphasize their surface accessibility, which is presumably important for the mechanism of regulation by PAE2072.