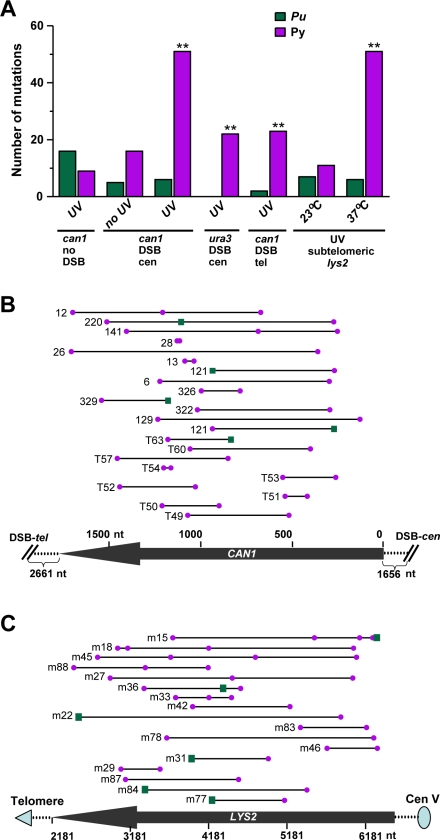
Figure 6. UV-induced and spontaneous mutants associated with DSB-repair and uncapped telomere arrest.
(A) Nucleotide changes in simple base substitutions associated with single and multiple mutant alleles. Bars represent the numbers of simple base substitutions in purine (Pu) or pyrimidine (Py) nucleotides in the unresected strand or in the coding strand, if no resection was expected. Detailed information about DNA sequences and mutation types in all mutant alleles is provided in Tables S4, S5, S6, S7, S8, S9, S10, S11, and S12 and summarized in Tables S16, S17, and S18. All sequenced mutants were from the wild-type (for can1 mutants) or cdc13-1 (for lys2 mutants) strains. All simple base substitutions from can1 mutants induced by 20 and 45 J/m2 in the DSB-cen strains were pooled. Other categories are the same as in Table 1 and in Tables S16, S17, and S18. Statistically significant differences in comparison to the “no-DSB-UV” controls for can1 mutants and to the “no arrest (23°C)” control for subtelomeric lys2 mutants are indicated by “**”. (B, C) Simple base substitutions in multiple mutant alleles. Mutations of CAN1 (B) are summarized from Tables S6, S7, and S8 and S10 and mutations of LYS2 (C) from Table S12. Multiple mutant alleles are shown as lines connecting individual mutations. Substitutions in pyrimidines (purple balls) and substitutions in purines (green squares) are shown above their positions in either CAN1 or LYS2 ORF (bottom lines with the arrowheads). Base substitutions are shown for the unresected strand. Indels and complex mutations are not shown. For the subtelomeric LYS2 reporter only distances from telomere are shown. Since presented can1 mutant alleles were obtained in strains with a DSB-site placed either on telomeric (allele numbers with a prefix “T”) or on centromeric side (numbered without a prefix) of the CAN1, coordinates are given within the ORF. Also shown are the distances of the ORF ends from a break in each type of constructs (DSB-tel or DSB-cen).