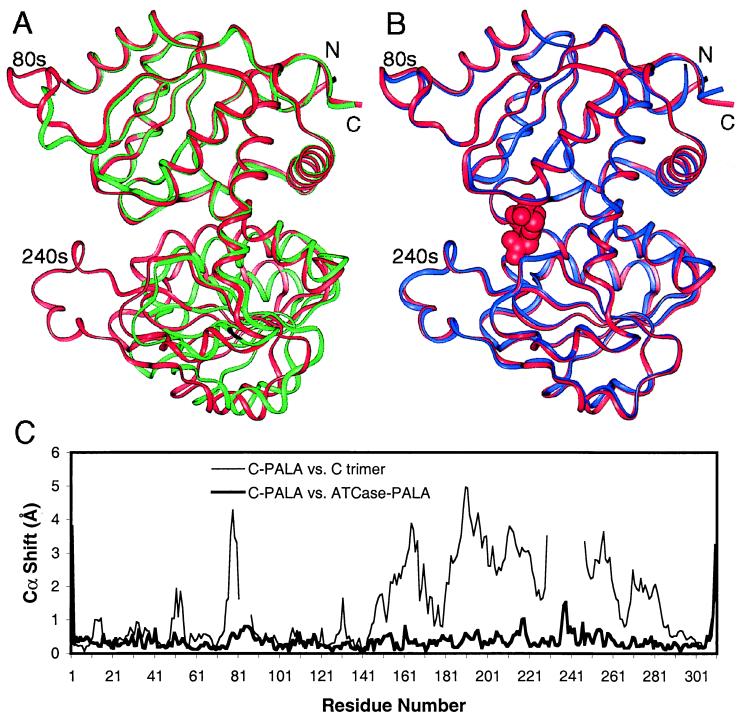
Figure 2.
The catalytic chains in the C trimer exhibit pronounced hinge closure upon PALA binding and adopt a conformation very similar to that of the chains in the PALA-bound holoenzyme. (A) Superposition of the N-terminal domains of catalytic chains from the C trimer–PALA complex (red) and the unliganded C trimer (PDB ID 3csu, Y chain, green). The chains superimpose closely within each domain but exhibit a 9.5° difference in hinge angle. (B) Superposition of catalytic chains from the C trimer–PALA complex (red) and the holoenzyme–PALA complex (PDB ID 1D09, blue). The similarity of the liganded conformations indicates that PALA binding is sufficient to promote the closed hinge in the holoenzyme. PALA is shown in red as a space-filling model. (C) Shift plots depicting differences in Cα positions as a function of residue number. Shift corresponding to A (chain from the C trimer–PALA complex versus the Y chain of the unliganded C trimer) is shown as a thin line illustrating a large relative motion of one domain with a hinge near Leu-140. Shift plot corresponding to B (chain from the C trimer–PALA complex versus a catalytic chain from the holoenzyme–PALA complex) is shown as a thick line. This comparison shows close similarity, except at the termini and the 240s loop.