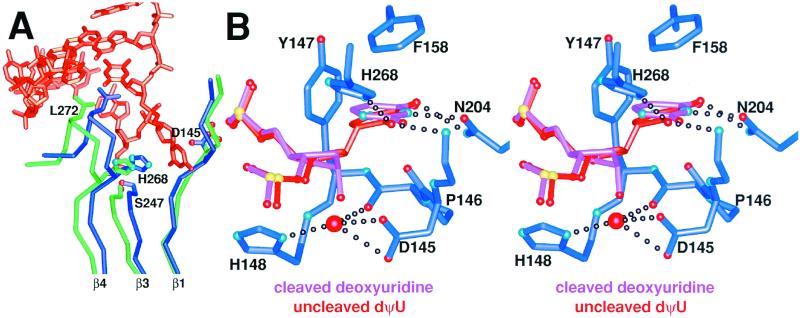
Figure 3.
A UDG global conformational change on binding substrate DNA creates the enzyme active center, which thereafter remains unchanged during the glycosylic bond cleavage reaction. (A) Superposition of apo-UDG (green) and DNA-bound UDG (dark blue) with the uncleaved substrate DNA (orange) shows that UDG undergoes an architecturally determined conformational closing on binding substrate DNA. β1 and β3 increase the number of interstrand hydrogen bonds between them and thereby zip up the UDG β-zipper. This creates the catalytically competent active site by bringing L272 into the DNA base stack and H268 and D145 into the active center. (B) Superposition of the uncleaved-substrate (orange) and product (pink) UDG–DNA complexes reveals the basis of glycosylic bond cleavage by UDG. As the conformation of UDG (dark blue carbon tubes) remains unchanged throughout the reaction (see text), only the protein conformation from the dΨU structure is shown.