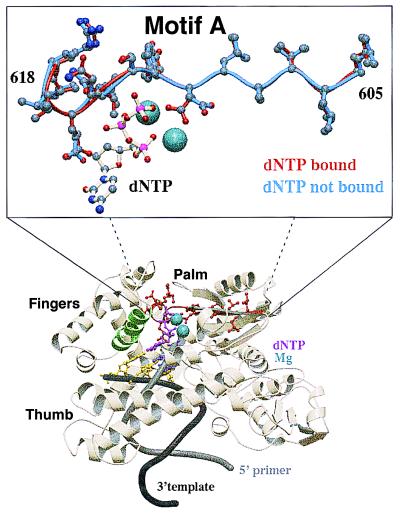
Figure 1.
Structure of Taq pol I bound with DNA and incoming dNTP. Evolutionarily conserved motif A (amino acids 605 to 617 highlighted in red) is located within the heart of the polymerase catalytic site. Residues of motif A interact with the incoming dNTP and amino acids in the finger motif during the conformational change step, subsequent to nucleotide binding. Motif A is superimposable in all polymerases with solved structures and begins at a hydrophobic antiparallel β-sheet that proceeds to an α-helix. The orientation of side chains within amino acids of motif A is nearly identical before (in blue) and subsequent to (in red) dNTP binding, with the exception of Asp-610, which rotates around the β carbon while coordinating with the Mg2+–dNTP complex. Motif B (amino acids 656 to 670) is highlighted in green. Coordinate sets 3ktq (ternary complex, closed form) and 4ktq (binary complex) from ref. 4 were obtained from the Protein Data Bank. The coordinates were aligned by using the 1sq_expl command in the software o to generate the inset view.