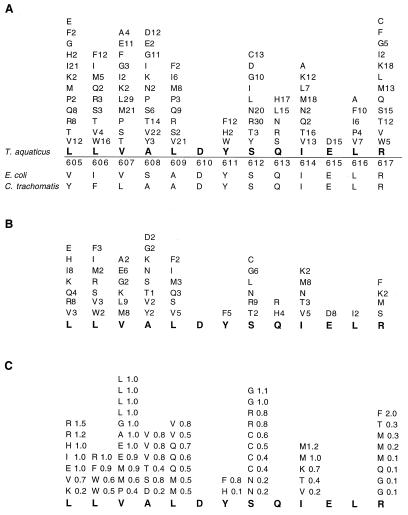
Figure 3.
High mutability of motif A. To test the importance of this conservation, Residues L605 to R617 (D610YSQIELR617) were randomly mutated such that each contiguous amino acid can be replaced by potentially any of the other 19. (A) The degree of mutability of each amino acid within motif A from all active clones (>10% to 200% activity relative to WT) complementing an E. coli DNA polymerase I temperature-sensitive strain. Amino acid substitutions at the locus are listed, along with the number of times each substitution is observed. (B) Mutations in clones exhibiting high activity (66% to 200% WT). (C) Mutations in clones containing a single amino acid substitution followed by activity relative to WT. Clones containing the same amino acid substitution contain unique silent changes at other loci; thus, they represent independent sequences.