FIGURE 3.
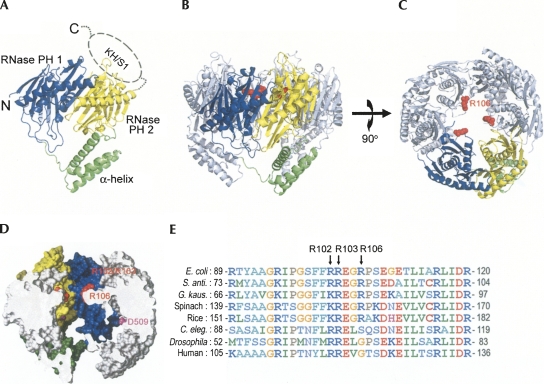
Crystal structure of E. coli. PNPase. (A) The structure of a monomeric PNPase contains three domains: the RNase PH1 domain (blue), the α-helix linker domain (green), and the RNase PH2 domain (yellow). The C-terminal KH/S1 domain (marked by dashed circle) was disordered and thus not visible in the crystal structure. (B) The side view of the trimeric PNPase with the only one of the monomers colored in blue, yellow, and green. The other two monomers are colored in gray. The side chain of R106 is displayed as a red sphere model. (C) The top view of the trimiric PNPase. The homotrimeric PNPase is assembled into a ring-like structure with a central channel. (D) The side view of the halved PNPase trimer shows the shape of the hollow central channel. The molecular surface of RNase PH1 domain is displayed in yellow, whereas the surface of RNase PH2 domain is displayed in blue. The channel has two constricted necks formed by arginine residues in the first RNase PH domain, R102 and R103 in the upper neck closer to the channel entrance, and R106 in the lower neck closer to the active site. The D509 residue marks the putative RNase active site located in the second RNase PH domain. (D) Sequence alignment of PNPase from different species shows that the basic arginine residues, R102, R103, and R106, located at the neck regions in the central channel are highly conserved in bacteria, plants, and animals. Sequences included are from Escherichia coli, Streptomyces antibioticus, Geobacillus kaustophilus, Spinacia oleracea (spinach), Oryza sativa (rice), Caenorhabditis elegans, Drosophila melanogaster, and human. The alignment was generated by ClustalW (http://www.ch.embnet.org/software/ClustalW.html).