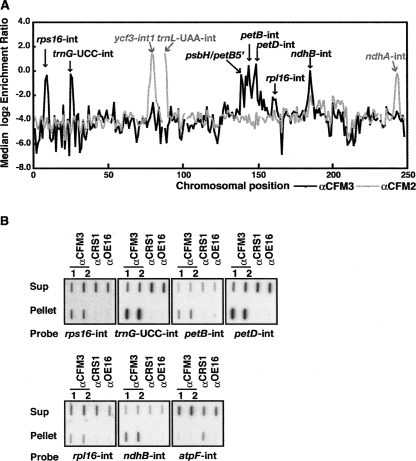
FIGURE 3.
Identification of chloroplast RNA ligands of ZmCFM3 in coimmunoprecipitation assays. (A) Summary of RIP-Chip data demonstrating the different populations of chloroplast RNAs that associate with CFM3 and CFM2. The median log2 transformed enrichment ratios (F635/F532, representing signal in the immunoprecipitation pellet/supernatant), are plotted according to chromosomal position for three replicate CFM3 RIP-Chip assays (black line). The RIP-Chip results reported previously for CFM2 (Asakura and Barkan 2007) are shown for comparison (gray line). Fragments for which fewer than two replicate spots per array yielded an F532 signal above background were excluded and appear as gaps in the curve. (B) Validation of RIP-Chip data by slot-blot hybridization. RNA purified from the pellets and supernatants of immunoprecipitations with antisera to the indicated proteins was applied to slot blots and hybridized to intron-specific probes. Antisera from different rabbits immunized with the CFM3 antigen were used in replicate assays (αCFM3 1 and 2). An immunoprecipitation with antibody to OE16, a protein in the thylakoid lumen that does not bind RNA, was used as a negative control. CRS1 is an atpF-specific chloroplast splicing factor and was analyzed as an additional control. One-third of the RNA recovered from each immunoprecipitation pellet and one-sixth of the RNA recovered from each supernatant was applied to each slot.