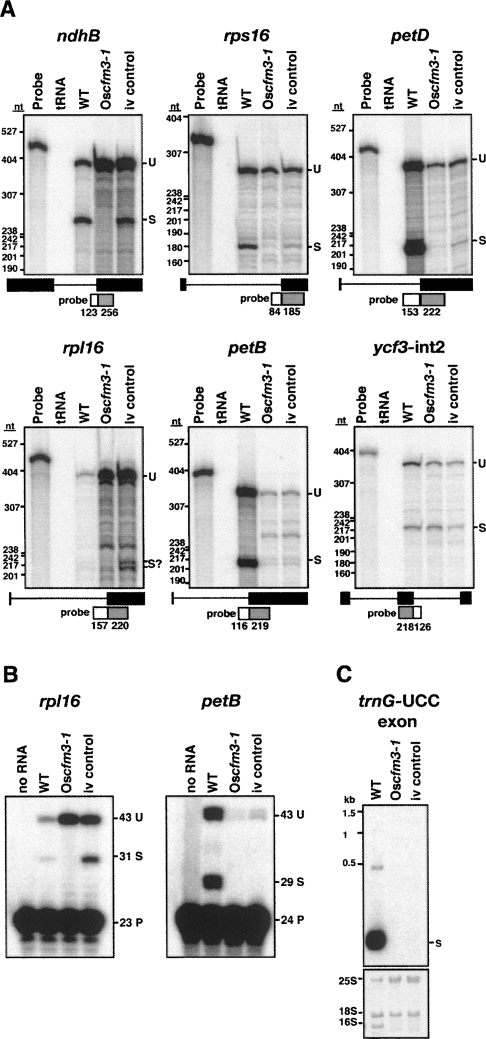
FIGURE 8.
Chloroplast splicing defects in the Oscfm3-1 mutant. RNA was analyzed from a normal seedling (WT), a Oscfm3-1 homozygote, and from an OsCFM3+ albino mutant segregating in the same background (iv control). (A) Ribonuclease protection assays. Total leaf RNA (2 μg) was analyzed using probes spanning either the 3′ or 5′ splice junction, as diagrammed. The length in nucleotides of probe segments corresponding to intron and exon sequences are shown in the diagrams. The positions of DNA size standards are shown to the left. U, unspliced; S, spliced. (B) Poisoned-primer extension assays. Total leaf RNA (4 μg) was used in reverse-transcription reactions using primers mapping several nucleotides downstream of the indicated intron. A dideoxynucleotide present in each reaction terminates reverse-transcription after different distances on spliced and unspliced RNA templates. The length in nucleotides of each product is indicated. U, unspliced; S, spliced; P, primer. (C) RNA gel blot showing loss of trnG-UCC in Oscfm3-1 mutant. Total leaf RNA (2 μg) was probed with a maize trnG-UCC exon probe. The same blot stained with methylene blue is shown in below.