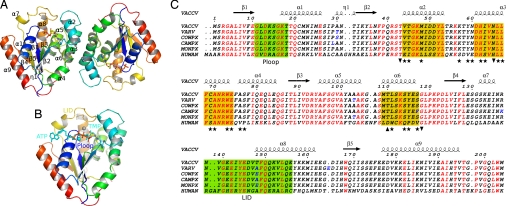
Fig. 1.
Quaternary and primary structures of vaccinia virus TMPK. (A) Overall structure of the TMPK dimer. The cartoon of the Vacc-TMPK dimer as seen in the asymmetric unit, rainbow colored with ligands shown as sticks and secondary structure numbering on subunit A. (B) Superimposition of the nucleotides from the human enzyme (PDB ID 1e2q) at the active site of Vacc-TMK chain B. ATP, TMP and magnesium from hTMPK are shown in cyan, whereas TDP and PPi and Mg2+ from Vacc-TMPK are colored according to atom type. (C) Sequence alignment of human, vaccinia (VACCV), variola (VARV), cowpox (COWPX), camelpox (CAMPX), and monkeypox (MONPX) viruses TMP kinases. Identical residues are in red. The TMP kinases of the pox family viruses are identical except for a few conservative substitutions (residues in blue), mostly in loops. Secondary structure elements from the Vacc-TMPK crystal structure are shown above the alignment. The P-loop and LID are boxed in green. Helices involved in dimer contacts are boxed in yellow. The positions of residues involved in the dimer interface of both enzymes are marked with stars, and those unique to human or vaccinia enzyme are marked as up and down triangles, respectively. This figure was created by using ESPript (51).