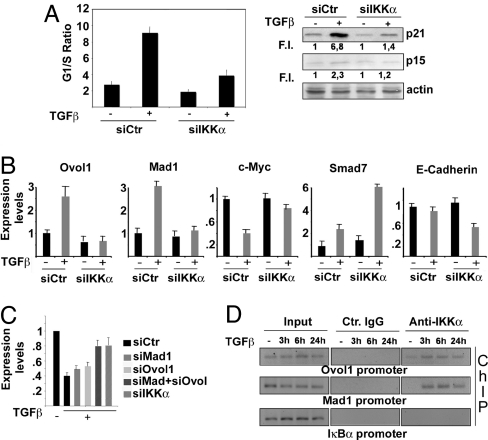
Fig. 3.
IKKα is part of the TGF-β-activated antiproliferative pathway in human keratinocytes. (A) HaCaT cells were transfected with either control or IKKα-specific siRNA. The transfected cells were treated with TGF-β for 24 h or left untreated and stained with propidium iodide to determine cell cycle profile. Results from 3 independent experiments performed in duplicate are expressed as G1/S ratio. Parallel samples were used to detect p21 and p15-INKB expression in total cell lysates. Numbers under each lane indicate fold-induction relative to controls. Band intensities were quantified by Kodak image software. (B) HaCaT cells were transfected with either control or IKKα siRNAs and treated with TGF-β as described above. RT-qPCR was performed using primers specific to the indicated genes. Results are mean expression values of 3 different experiments performed in triplicate. Error bars are standard deviations. (C) HaCaT cells were transfected with either control siRNA or siRNAs specific for Mad1, Ovol1, Mad1 plus Ovol1, or IKKα, and the ability of TGF-β to down-regulate c-Myc was evaluated by RT-qPCR. Results are the mean expression values of 3 different experiments performed in triplicate. Error bars indicate standard deviation. Expression values in untreated cells were set as 1. (D) HaCaT cells were treated with TGF-β for the indicated times. IKKα-containing protein–DNA complexes were immunoprecipitated with IKKα-specific antibody. PCR was performed on the recovered DNA using specific primers for a region spanning positions −1337 to −1170 of the Mad1 gene and −747 to −555 of the Ovol1 gene. Control IgG and PCR on IkBα promoter served as negative controls. PCR primers were designed based on location of potential TGF-β-responsive elements (SMAD-binding sites) identified by MatInspectorSoftware (Genomatix GmbH).