Figure 3. Interaction of dCTCF with gypsy insulator proteins.
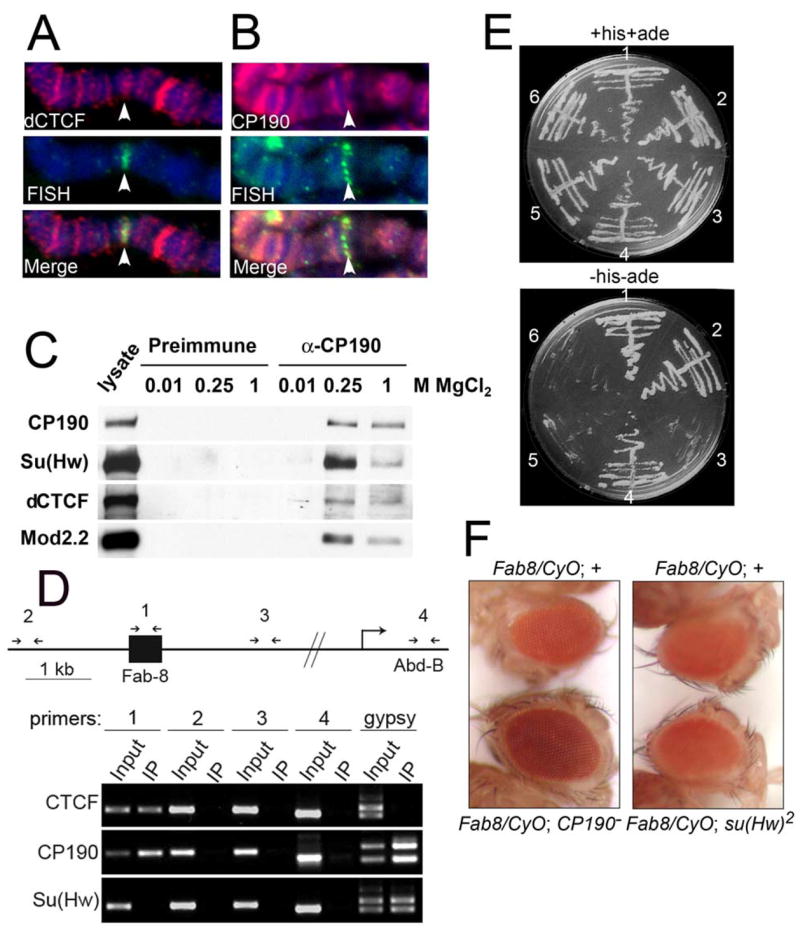
(A) dCTCF is present at the cytological location of the Fab-8 insulator. Red indicates the immunolocalization of dCTCF, green indicates the localization of Fab-8 sequences by FISH and DNA is stained with DAPI in blue. Arrow points to the location of Fab-8.
(B) CP190 is present at the cytological location of the Fab-8 insulator. Red indicates the immunolocalization of CP190, green indicates the localization of Fab-8 sequences by FISH and DNA is stained with DAPI in blue. Arrow points to the location of Fab-8.
(C) Western analysis of control preimmune (lanes 2–4) and α-CP190 column (lanes 5–7) immunoaffinity purifications to verify the presence of CP190, Su(Hw), dCTCF and Mod(mdg4)2.2 in α-CP190 eluates at the indicated concentrations of MgCl2.
(D) Chromatin immunoprecipitation analysis of the Fab-8 insulator. The top part of the panel is a diagram of the 5′ region of the Abd-B gene, including the Fab-8 insulator. Numbered arrows above the diagram indicate the location of primers used in the PCR reactions. The lower portion of the panel shows the result of the electropheretic analysis of the PCR products after immunoprecipitation with antibodies to the proteins indicated on the left. For each primer pair, an input and IP lane are shown. Also shown is a control using primers to the insulator present in the gypsy retrotransposon.
(E) Growth of yeast strains expressing dCTCF and Su(Hw), CP190 or control on nonselective (+histidine +adenine) (top) or selective ( histidine -adenine) (bottom) media. Sector 1, Su(Hw)AD-CP190BD. Sector 2, dCTCFAD-CP190BD. Sector3, dCTCFAD-CP190-BTB domainBD. Sector 4, dCTCFAD-CP190-carboxytermBD. Sector 5, dCTCFAD-emptyBD. Sector 6, CP190BD-emptyAD.
(F) Left panel: Eyes of male flies of the genotype Fab-860.39.2/CyO;+/+ (top) and Fab-860.39.2/CyO; CP1904-1/CP190H31-2 (bottom). Right panel: Eyes of male flies of the genotype Fab-860.39.2/CyO;+/+ (top) and Fab-860.39.2/CyO; su(Hw)2 (bottom).
