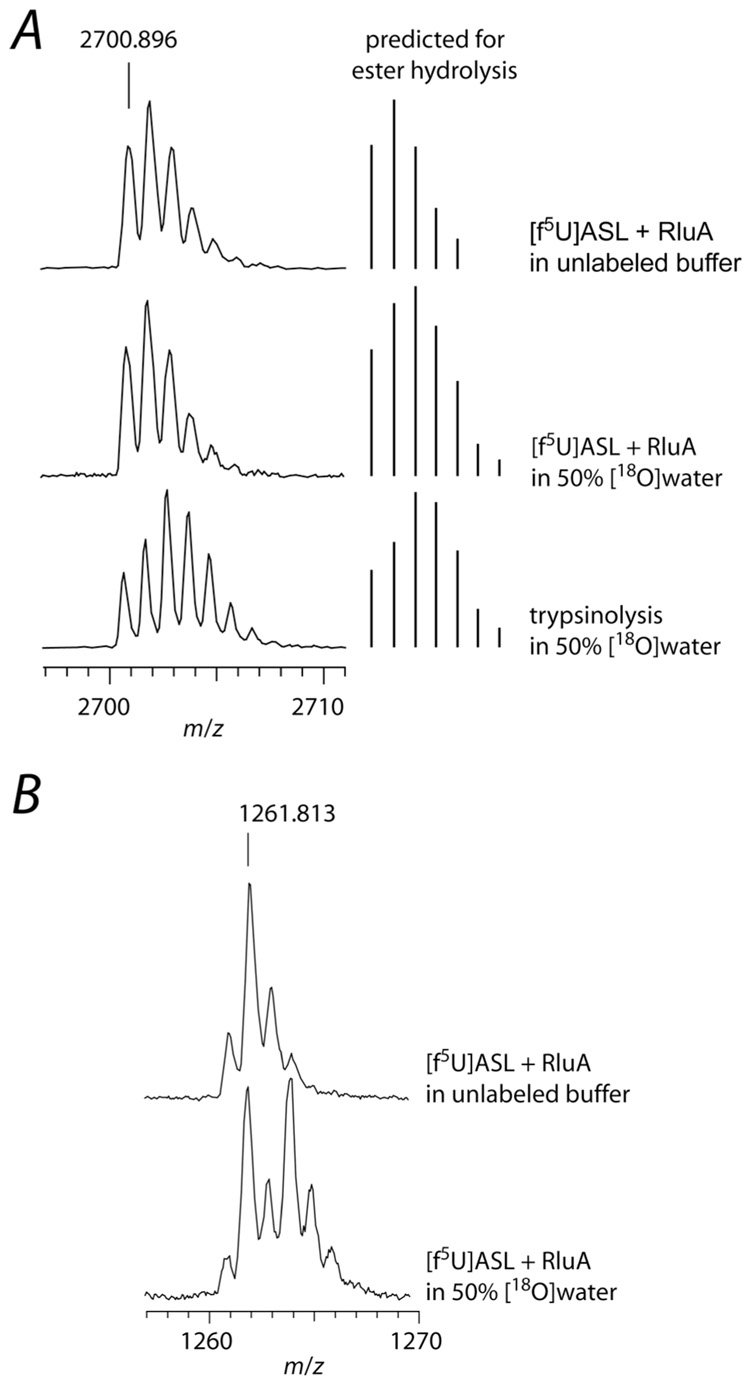
Figure 8.
Labeling studies to probe for ester hydrolysis during the decomposition of the adduct between RluA and [f5U]ASL. The adduct was formed with a 2-fold excess of RluA and then heat-disrupted, and the protein and RNA were separately digested and subjected to MALDI-MS analysis. A, Partial mass spectra of the tryptic digest of RluA showing the tryptic peptide 53Leu-Lys77 (2700.4 m/z predicted) containing Asp-64 (left) and the predicted mass distribution for ester hydrolysis of a Michael adduct (right); top, RluA incubated in unlabeled buffer with [f5U]ASL; middle, RluA incubated with [f5U]ASL in buffer containing [18O]water (50%), showing no 18O incorporation into the peptide, which does not match the prediction for ester hydrolysis; bottom, RluA incubated in unlabeled buffer with [f5U]ASL with trypsinolysis in buffer containing [18O]water (50%). B, Partial mass spectra of [f5U]ASL after incubation with RluA and digestion with RNase T1; top, incubation in unlabeled buffer; bottom, incubation in buffer containing [18O]water (50%), showing 18O incorporation into the hydrated [f5U]ASL (50% of the product contains 16O and 50% contains 18O and is therefore shifted +2 m/z). The small [M]+ peak observed could be due to amination rather than hydration since ammonium chloride (100 mM) was present in the reaction mixture; however, the [M]+ was still present when ammonium chloride was replaced with sodium chloride (data not shown), so it more likely arises from the high laser power necessary to obtain the mass spectra of the oligonucleotides.