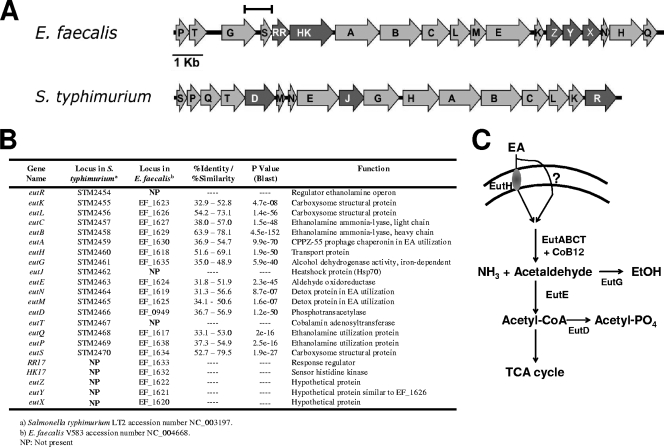
FIG. 1.
eut operon for EA utilization in E. faecalis and Salmonella enterica serovar Typhimurium (S. typhimurium). (A) A diagram of the gene organization in the two operons is shown with the darker shade of gray representing genes that are present in one organism but missing in the other. The bar represents the extent of the fragment cloned in the pTCV-RR17p transcriptional fusion plasmid. (B) Table summarizing gene names, annotation numbers in genome databases, the results of ClustalW and BLAST analyses, and function. (C) Schematic representation of the basic EA metabolism in Salmonella. EA can enter the cell by diffusion either via EutH or by other, unknown routes (38). EutBC are the two subunits of the cobalamin-dependent EA ammonia-lyase. EutE is an acetaldehyde dehydrogenase, EutG is an alcohol dehydrogenase, and EutD is a phosphotransacetylase (6, 27, 39). Notably, the genes for the tricarboxylic acid cycle (TCA) are absent in the genome of E. faecalis V583.