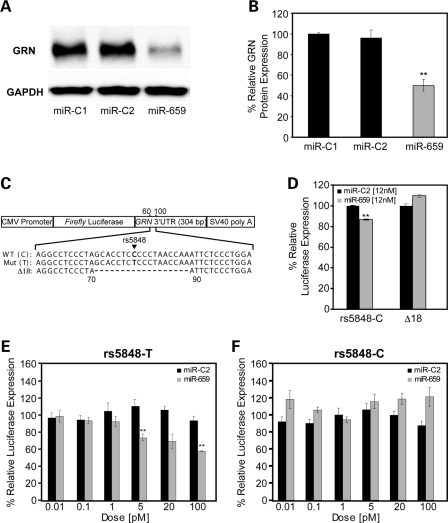
Figure 4.
Functional analyses of the GRN regulation by miR-659 in vitro. (A) Representative immunoblot analyses of human M17 cells transfected with miR-659 or control miRNAs (miR-C1 and miR-C2). (B) Quantification of immunoblotted GRN in human M17 cells transfected with miR-659 or control miRNAs. GRN expression was normalized to GAPDH. Data are from four independent experiments and represent mean ± SEM. (**indicates P < 0.001; two sample t-test). (C) Schematic overview of the pMIR-REPORT vector containing a CMV promoter, firefly luciferase gene, full-length 3′-UTR of GRN and SV40 poly A-tail. Constructs with wild-type C-allele and risk T-allele of rs5848 at position 78 in 3′-UTR were created as well as a Δ18 construct in which the complete predicted binding site of miR-659 was deleted (position 71–88 in the 3′-UTR). (D) Luciferase expression in N2A cells transfected with pMIR-REPORT-rs5848C or pMIR-REPORT- Δ18 and co-transfected with high dose (12 nm) miR-659 or miR-C2. Relative luciferase activity was determined as firefly luciferase activity normalized to Renilla luciferase activity. Each experiment was repeated six times. miR-659 significantly decreased the expression of firefly luciferase using the wild-type pMIR-REPORT-rs5848C vector, but not using the pMIR-REPORT-Δ18 in which the miR-659 binding site was deleted. Data represent mean ± SEM. (**indicates P < 0.001; two sample t-test). (E–F) pMIR-REPORT-rs5848C and pMIR-REPORT-rs5848T vectors were transfected in N2A cells and co-transfected with variable low doses (0.01–100 pM) of miR-659 or miR-C2. Each experiment was repeated three times. A significant reduction in the expression of firefly luciferase was observed in the presence of 5 and 100 pM of miR-659 from the rs5848T construct (D, **P < 0.02; two-tailed t-test), while no reduction in the expression of firefly luciferase from the rs5848C construct was observed at any of the doses. Data represent mean ± SEM.