Abstract
Hereditary nonpolyposis colorectal cancer is caused by germline mutations in DNA mismatch repair genes. The majority of cases are associated with mutations in hMSH2 or hMLH1; however, about 12% of cases are associated with alterations in hMSH6. The hMSH6 protein forms a heterodimer with hMSH2 that is capable of recognizing a DNA mismatch. The heterodimer then utilizes its adenosine nucleotide processing ability in an, as of yet, unclear mechanism to facilitate communication between the mismatch and a distant strand discrimination site. The majority of reported mutations in hMSH6 are deletions or truncations that entirely eliminate the function of the protein; however, nearly a third of the reported variations are missense mutations whose functional significance is unclear. We analyzed seven cancer-associated single amino acid alterations in hMSH6 distributed throughout the functional domains of the protein to determine their effect on the biochemical activity of the hMSH2-hMSH6 heterodimer. Five alterations affect mismatch-stimulated ATP hydrolysis activity providing functional evidence that missense variants of hMSH6 can disrupt mismatch repair function and may contribute to disease. Of the five mutants that affect mismatch-stimulated ATP hydrolysis, only two (R976H and H1248D) affect mismatch recognition. Thus, three of the mutants (G566R, V878A, and D803G) appear to uncouple the mismatch binding and ATP hydrolysis activities of the heterodimer. We also demonstrate that these three mutations alter ATP-dependent conformation changes of hMSH2-hMSH6, suggesting that cancer-associated mutations in hMSH6 can disrupt the intramolecular signaling that coordinates mismatch binding with adenosine nucleotide processing.
The process of DNA mismatch repair (MMR)2 is essential for maintaining genomic stability through recognition and correction of mismatched nucleotides and small insertion/deletion loops. In human cells, three MutS homologs (hMSH2, hMSH3, and hMSH6) assemble into two functional heterodimers that recognize a DNA lesion and initiate the MMR process. The hMSH2-hMSH6 heterodimer is most abundant and recognizes single base mismatches and small insertion/deletion loops. It signals for repair through a heterodimer of MutL homologs, hMLH1-hPMS2. In addition to initiation of repair, these MMR proteins also participate in signaling for cell cycle arrest and apoptosis in response to DNA damage (1, 2).
Key conserved features of all MutS homologs, including hMSH2 and hMSH6, are highly conserved ATP-binding and hydrolysis motifs located near the C termini (3). Their ability to bind and hydrolyze ATP is crucial for MMR function, and this ATP hydrolysis (ATPase) activity is stimulated by a DNA mismatch (3–6). DNA binding by MSH2-MSH6 occurs in the presence of ADP and is inhibited by ATP (4, 7–9). Mismatch binding leads to a rapid exchange of ADP for ATP resulting in a conformation change to a sliding clamp that freely diffuses along DNA. When the ends of the DNA molecule are blockaded, the MSH2-MSH6 sliding clamps remain trapped on the DNA (9, 10). This conformational change and movement on the DNA requires ATP but not ATP hydrolysis (9, 10). These observations led to the formation of a “molecular switch” model for hMSH2-hMSH6 function, which proposes that multiple hMSH2-hMSH6 complexes load at the site of a DNA mismatch, and through an ATP-dependent passive diffusion along the DNA, they communicate a signal between the mismatch and a distant strand discrimination signal (4, 11).
Cells lacking functional MMR accumulate mutations at an increased rate (12). This mutator phenotype is underlined by the causal role that loss of MMR plays in the heritable cancer syndrome, hereditary non-polyposis colorectal cancer (HNPCC) (13). Although germline mutations in hMSH2 or hMLH1 account for the majority of HNPCC cases, a subset (12%) of reported MMR mutations are found in hMSH6 (MMR Genes Variant data base) (14). Tumors with mutated hMSH6 display a later age of onset and are less likely to fulfill the Amsterdam criteria for HNPCC (15) than tumors with hMSH2 mutations. Interestingly, female hMSH6 mutation carriers have a higher frequency of endometrial cancers and endometrial hyperplasia (16, 17). These differences in clinical characteristics make understanding the underlying biochemical defects associated with mutations of hMSH6 important for elucidating its role as a tumor suppressor.
Although most cancer-associated mutations in hMSH6 result in truncations or deletions of the protein, about one-third are missense mutations encoding a single amino acid change (14). The biochemical defects associated with these missense mutations are unknown. In this study, we have analyzed seven missense mutations in hMSH6 that were reported in HNPCC and suspected HNPCC families (18–20). We analyzed the recombinant mutant proteins in conjunction with their wild-type hMSH2 partner for DNA binding and adenosine nucleotide processing. We found that five of the mutations resulted in decreased in vitro biochemical functions thereby supporting their roles in tumorigenesis.
EXPERIMENTAL PROCEDURES
hMSH6 Mutant-containing Heterodimer Preparation—hMSH6 missense mutants were generated by QuickChange site-directed mutagenesis (Stratagene). Wild-type and mutant hMSH2-hMSH6 heterodimers were expressed and purified as described previously (21). Protein concentration was determined by absorbance at 280 nm. For some mutants, dimer stability was examined by observing denaturation in the presence of urea. 1 μg of protein was incubated with 0–7 m urea in buffer containing 25 mm Hepes (pH 8.1), 100 mm NaCl, and 5.3% glycerol for 15 min at room temperature. The reactions were analyzed on a 5% native gel and silver-stained to visualize proteins. A separate gel was run for Western blot analysis using polyclonal hMSH2 (Calbiochem) and hMSH6 (Bethyl Laboratories) antibodies to identify bands corresponding to hMSH2-hMSH6 complex and monomer forms. The monomer and dimer signal intensity was quantified using ImageJ (22).
ATPase Assays—ATPase activity was measured at multiple concentrations (15–100 nm) of hMSH2-hMSH6 protein by incubating with 16.5 nm [γ-32P]ATP (PerkinElmer Life Sciences) in 25 mm Hepes (pH 8.1), 100 mm NaCl, 10 mm MgCl2, 1 mm DTT, 15% glycerol, and 20–240 μm unlabeled ATP in 20-μl reactions at 37 °C for 30 min. Reactions were carried out in the presence of 41-bp DNA oligonucleotide containing a central G/T mismatch. The fraction of hydrolyzed [γ-32P]ATP was determined by charcoal binding as described previously (4). For analysis of DNA-independent ATPase activity, protein concentration was increased to 200 nm.
Surface Plasmon Resonance (SPR) Analysis—The Biacore T100 (GE Healthcare) optical biosensor was used to monitor real time interaction of hMSH2-hMSH6 with DNA. A 41-bp biotinylated DNA oligonucleotide containing a central G/T mismatch was affixed to a streptavidin sensor chip (GE Healthcare) to ∼100 resonance units. 0–180 nm of hMSH2-hMSH6 were injected over the chip at a flow rate of 30 μl/min for 3 min in buffer containing 25 mm Hepes (pH 8.1), 110 mm NaCl, 1 mm DTT, 2 mm MgCl2, 2% glycerol, and 25 μm ADP. After 3 min, protein flow was halted, and injection of buffer alone was continued for another 3 min to observe dissociation. Samples were kept at 10 °C prior to injection, and experiments were performed at 25 °C. The DNA chip surface was regenerated after each run by washing with 2.5 m NaCl at 100 μl/min for 1 min. Data were analyzed by fitting each binding curve to a 1:1 binding model using Biacore T100 analysis software (GE Healthcare). ATP-dependent dissociation was observed using SPR by pre-binding 50 nm hMSH2-hMSH6 to heteroduplex DNA for 3 min as above. During the dissociation phase, protein flow was halted, and an identical buffer with 250 μm ATP was injected over the surface at 30 μl/min. Dissociation of the protein-DNA complex was observed for 3 min.
Adenosine Nucleotide Binding and Exchange—ATP crosslinking experiments were performed with hMSH2-hMSH6 protein (150 nm) incubated with 0.5 μm [γ-32P]ATP for 20 min at room temperature in 20 μl of buffer containing 25 mm Hepes (pH 8.1), 100 mm NaCl, 1 mm DTT, 11% glycerol, and 0 or 5 mm MgCl2 as indicated. The reactions were cross-linked on ice for 10 min using a UV Stratalinker 1800 (Stratagene) and separated on a 6% SDS-polyacrylamide gel. Bands were visualized using the Cyclone Storage Phosphor System (PerkinElmer Life Sciences) and OptiQuant software (Packard Instrument Co.). 0.7 μg of each protein was run on a separate gel and stained with Coomassie Blue to control for the amount of mutant protein loaded. Signal intensity was determined using ImageJ (22). ATPγS filter binding experiments were performed with 100 nm hMSH2-hMSH6 protein, 0.05 μm [35S]ATPγS (PerkinElmer Life Sciences), and 0.5–20 μm unlabeled ATPγS as described previously (21). [35S]ATPγS bound was measured in a Beckman LS 6500 counter. ADP → ATP exchange assays were performed with 40 nm hMSH2-hMSH6 incubated with 2.3 μm [3H]ADP (PerkinElmer Life Sciences) as described previously (21). 60 nm heteroduplex DNA and 0.5 mm ATP were added to start the reaction. Bound [3H]ADP was quantified using a Beckman LS 6500 counter.
Partial Trypsin Proteolysis—1 μg of hMSH2-hMSH6 protein was incubated in trypsin proteolysis buffer (50 mm ammonium bicarbonate, 25 mm Hepes, 100 mm NaCl, 1 mm DTT, 1 mm MgCl2, 0.1 mm EDTA, and 20% glycerol) with increasing amounts of trypsin (0–160 ng) in 20-μl reactions for 45 min. Reactions were stopped by the addition of Laemmli sample buffer and boiling. 0.5 μg of protein were loaded onto 8% SDS-PAGE and visualized by silver staining. Western blot analysis was performed to identify hMSH2 and hMSH6 fragments. Mass spectrometry was also used to identify peptide fragments in specific bands of interest. The bands were excised, and gel pieces were subjected to in-gel reduction, carboxyamidomethylation, and tryptic digestion (Sigma). As described previously (23), peptide sequences were determined by microcapillary reverse-phase chromatography directly coupled to a Finnigan LTQ quadrupole ion trap mass spectrometer equipped with a commercial nanoelectrospray source. Interpretation of the resulting tandem mass spectroscopy spectra of the peptides was facilitated by programs developed in the University of Connecticut Health Center Proteomic and Biological Mass Spectrometry facility and by data base correlation with the algorithm Sequest (24).
RESULTS
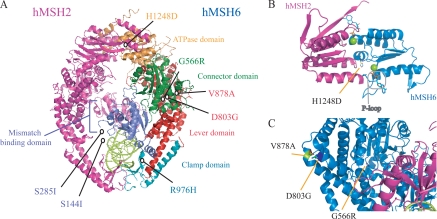
HNPCC-associated hMSH6 Missense Mutations Do Not Affect Ability of the Protein to Interact with hMSH2—We examined seven cancer-associated missense mutations in hMSH6 identified in HNPCC kindreds or familial cancer patients suspected of having HNPCC (18–20) to determine their effects on the biochemical activity of the hMSH2-hMSH6 heterodimer. These mutations encode for single amino acid substitutions (S144I, S285I, G566R, D803G, V878A, R976H, and H1248D), which lie throughout the structural subdomains of hMSH6 as determined from the hMSH2-hMSH6 crystal structure (Fig. 1A) (25). The hMSH2-hMSH6 heterodimer was purified via a hexahistadine tag on the N terminus of hMSH6. hMSH6 and hMSH2 co-purify in an apparent 1:1 ratio as determined by Coomassie staining of an SDS-polyacrylamide gel demonstrating that none of the missense mutations affect heterodimer formation (data not shown). We further examined the G566R and H1248D mutants for effects on heterodimer stability. The G566R mutation had been suggested previously to affect dimer stability when expressed in mammalian cell culture (26). The H1248D mutation is located near the C terminus of hMSH6 in a region that overlaps with hMSH2 (Fig. 1B) in a domain that had been determined to affect dimer interaction (27). Analysis of the dimer to monomer ratio following ureadependent denaturation does not reveal effects of either mutation on heterodimer stability (supplemental Fig. 1).
FIGURE 1.
Location of hMSH6 missense mutations. A, seven hMSH6 missense mutations tested mapped to the human hMSH2-hMSH6 heterodimer crystal structure (25). The hMSH2 subdomain is indicated in pink, and the hMSH6 subunit is labeled by subdomain. The missense mutations lie throughout the subdomains of hMSH6. S144I and S285I lie in an N-terminal region of the protein that was removed prior to crystallization. DNA containing a mispair is shown in lime green. B, two overlapping ABC ATPase active sites of hMSH2 (pink) and hMSH6 (blue) are formed at the C-terminal heterodimer interface. ADP is bound to the P-loop of each subunit. His-1248 of hMSH6 lies near the bound nucleotide on a parallel α-helix. C, reverse view of hMSH6 showing a close-up of the location of G566R, D803G, and V878A in the connector and lever domains. All structural representations were created with PyMOL.
Steady-state ATPase Activity Is Affected in HNPCC hMSH6 Missense Mutants—The MSH2-MSH6 heterodimer contains an intrinsic low level ATP hydrolysis activity that is stimulated in the presence of mismatched DNA and is essential for MMR (3–6). We determined the steady-state ATPase activity of hMSH6 missense mutants in the presence or absence of a DNA mismatch. In the absence of DNA, the majority of mutants showed only a slightly lower kcat than wild-type protein, with H1248D and V878A appearing more significantly affected (Table 1 and supplemental Fig. 2). The H1248D mutation is located in the ATPase domain (see Fig. 1B) in close proximity to where the γ-phosphate would reside and may coordinate with the ABC-ATPase signature helix of hMSH2 to promote hydrolysis of ATP in the hMSH6 nucleotide binding pocket. In the presence of heteroduplex DNA, more dramatic changes in kcat and catalytic efficiency (kcat/Km) were seen for five of the seven hMSH6 mutant proteins (Table 1 and supplemental Fig. 2). The G566R mutant, although similar in ATPase activity to wild-type protein in the absence of DNA, had a dramatic reduction in mismatch-stimulated ATPase, suggesting deficiencies in other aspects of its activity.
TABLE 1.
ATP hydrolysis activity of hMSH2-hMSH6 missense mutants in the presence and absence of a DNA mismatch In the presence of homoduplex DNA, wild-type kcat/Km is 19.7 × 10–4 m–1·min–1.
|
ATPase (G/T DNA)
|
ATPase (no DNA)
|
|||
|---|---|---|---|---|
| kcat | kcat/Km × 10–4 | kcat | kcat/Km × 10–4 | |
| min–1 | m–1·min–1 | min–1 | m–1·min–1 | |
| Wild type | 24.0 ± 4.2 | 35.8 | 1.6 ± 0.2 | 1.6 |
| S144I | 15.6 ± 1.3 | 36.0 | 1.1 ± 0.1 | 3.6 |
| S285I | 13.0 ± 1.0 | 38.3 | 0.8 ± 0.0 | 2.1 |
| G566R | 2.7 ± 0.2 | 12.6 | 1.2 ± 0.3 | 0.6 |
| D803G | 5.6 ± 0.4 | 9.7 | 0.6 ± 0.0 | 3.5 |
| V878A | 8.1 ± 0.8 | 26.2 | 0.5 ± 0.0 | 0.9 |
| R976H | 5.7 ± 0.5 | 30.8 | 1.3 ± 0.1 | 2.1 |
| H1248D | 1.0 ± 1.0 | 13.0 | 0.3 ± 0.0 | 0.6 |
DNA Mismatch Binding Is Not Affected for Most of the hMSH6 Mutants—Pre-steady-state kinetic experiments indicate that MSH2-MSH6 undergoes a rapid burst of ATP hydrolysis followed by a slow rate-limiting step (8), which may require mismatch binding to overcome (4). Thus, we sought to determine whether missense mutations in hMSH6 affect DNA mismatch recognition. The kinetics of hMSH2-hMSH6 binding to a 41-bp heteroduplex oligonucleotide was examined by SPR (supplemental Fig. 3). Multiple protein concentrations (0–120 nm) were analyzed to determine the equilibrium dissociation constant (KD(G/T)) (Table 2). All of the hMSH6 mutant-containing heterodimers retained an ability to interact with heteroduplex DNA, and most mutants displayed KD(G/T) similar to wild-type protein. Two mutants R976H and H1248D displayed reduced affinity for the mismatch with KD(G/T) values increased ∼4- and 7-fold, respectively. The R976H mutation lies in the clamp domain of the protein (Fig. 1A), and data from the crystal structure indicate that neighboring residues Asn-975 and Arg-974 interact with the DNA backbone during mismatch recognition (25); thus it is not surprising that this mutation would affect mismatch binding. However, as noted above, the H1248D mutation resides at the opposite end of the molecule in the ATPase domain suggesting that some mutations can have effects on functions that occur at a distant site in the molecule. A similar effect was observed with missense mutations in hMSH2 that affected mismatch binding, although far removed from the mismatch binding domain (21). The remaining hMSH6 mutations did not affect mismatch binding suggesting some mutations may uncouple mismatch recognition from mismatch-stimulated ATPase activity.
TABLE 2.
DNA and nucleotide binding properties and nucleotide exchange of hMSH2-hMSH6 mutants In the presence of homoduplex DNA, wild-type KD(G/C) is 48.6 nm.
| hMSH6 mutation | DNA binding, KD(G/T) | ATPγS binding, KD(ATP) | ADP-ATP exchange, t1/2 |
|---|---|---|---|
| nm | μm | s | |
| Wild type | 4.78 ± 0.70 | 0.91 ± 0.20 | 4.2 |
| S144I | 5.65 ± 0.19 | 1.10 ± 0.19 | 2.5 |
| S285I | 4.96 ± 0.07 | 0.37 ± 0.26 | 2.1 |
| G566R | 8.51 ± 1.00 | 4.19 ± 1.21 | 3.2 |
| D803G | 11.59 ± 0.97 | 0.41 ± 0.11 | 1.1 |
| V878A | 5.43 ± 0.52 | 1.27 ± 0.23 | 2.8 |
| R976H | 23.15 ± 2.88 | 0.52 ± 0.13 | 9.5 |
| H1248D | 39.79 ± 1.19 | 0.69 ± 0.23 | 4.9 |
Missense Mutations in hMSH6 Affect Binding to Adenosine Nucleotides—Despite the structural distance between the DNA binding and ATP hydrolysis domains, nucleotide binding has been shown to modulate the interaction of hMSH2-hMSH6 with DNA. ATP and ATPγS decrease protein affinity for DNA and induce a large conformational change in the protein allowing formation of a sliding clamp on DNA (9, 10). The two subunits MSH2 and MSH6 have differential specificity for ADP and ATP, with MSH6 having a higher affinity for ATP and MSH2 having a higher affinity for ADP (8, 28, 29). We examined the effect that hMSH6 missense mutations have on the interaction of hMSH2-hMSH6 with ADP, ATP, and ATPγS.
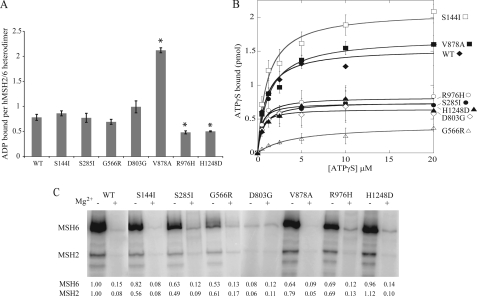
Wild-type hMSH2-hMSH6 and the S144I, S285I, G566R, and D803G each bind approximately one molecule of ADP per heterodimer (Fig. 2A). Three mutations in hMSH6 resulted in altered ability to bind ADP. R976H and H1248D had reduced binding ability compared with wild type. Interestingly, V878A was able to bind two molecules of ADP per heterodimer suggesting that both nucleotide states are occupied by ADP in this mutant. Although MSH2 has been demonstrated to have a higher affinity for ADP, the fact that mutations in hMSH6 could affect ADP binding may indicate a cross-talk between the subunits with regard to nucleotide binding as has been suggested previously (29).
FIGURE 2.
Adenosine nucleotide binding of WT and hMSH6 missense mutants. A, ADP binding activity. WT and mutant hMSH2-hMSH6 was incubated with [3H]ADP. The amount of ADP bound was determined by filter binding. Error bars indicate standard deviation of three trials. Differences in ADP binding to V878A, R976H, and H1248D were statistically significant (p < 0.01) compared with WT as indicated by asterisks. B, ATPγS binding. Varying concentrations of [35S]ATPγS were incubated with 100 nm hMSH2-hMSH6 protein. The amount of bound nucleotide was determined by filter binding. Points represent the average of three trials, and the data were fit with the Michaelis-Menten equation. C, ATPSγS binding activity. WT and mutant proteins were incubated and cross-linked to radiolabeled [γ-32P]ATP in the presence or absence of Mg2+ (to inhibit hydrolysis). Binding of ATP was visualized by phosphorimaging. Signal was adjusted for loading as measured by Coomassie stain. Values indicate quantitated band intensities for hMSH2 and hMSH6 normalized to WT-Mg2+.
We also used filter binding to examine the effect of hMSH6 mutations on ATPγS binding (Fig. 2B). Similar to our previously published study (21), the KD(ATPγS) for the wild-type heterodimer was 0.91 μm with slightly more than one molecule of ATPγS bound per heterodimer. The G566R mutant, however, was clearly deficient in ATPγS binding whereas lesser defects were observed for D803G, S285I, R976H, and H1248D. We then examined [γ-P32]ATP binding by UV crosslinking (Fig. 2C). These experiments were performed in the presence and absence of magnesium to prohibit hydrolysis of the γ-labeled phosphate. In the absence of magnesium, ATP is bound to both subunits and much more prominently by hMSH6, consistent with hMSH6 containing the high affinity site for ATP binding (8, 28, 29). As observed with the filter binding assays, the G566R mutant was affected in its ability to be crosslinked to ATP, although the most notable defect was observed in the D803G mutant heterodimer. The D803G mutation also affected the ability to cross-link ATP to the hMSH2 subunit, consistent with the idea of cross-communication between the subunits. More modest defects were observed for the S285I, V878A, and R976H mutants, although surprisingly, no defect was observed for the H1248D mutant. The H1248D mutant also showed similar reduction in ATP binding as wild type in the presence of magnesium, despite the dramatic defect in ATPase activity observed for this mutant. The reduced ATP hydrolysis function of this mutant might lead one to expect that the H1248D hMSH6 subunit would retain the γ-labeled phosphate even in the presence of magnesium, although this is not the case. This may suggest that the reduced γ-32P cross-linking to hMSH6 in the presence of magnesium is not because of hydrolysis of the labeled phosphate but rather is because of an inability of hMSH6 to bind ATP in the presence of magnesium. Taken together, the filter-binding and cross-linking studies reveal defects in ATP binding particularly for the G566R and D803G mutants that may partially explain their reduced mismatch-stimulated ATPase.
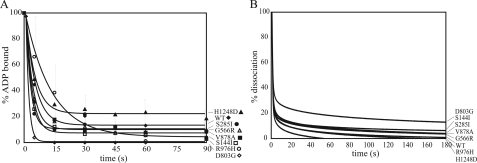
hMSH6 Missense Mutants Undergo ADP → ATP Exchange and Dissociation from a Mismatch—We measured the rate of mismatch-stimulated exchange of ADP for ATP for each mutant heterodimer by pre-binding the proteins to [H3]ADP and initiating the reaction with mismatched DNA and excess unlabeled ATP. Between 70 and 80% of the ADP is released in the first 5 s for both the wild-type and mutant heterodimers with the exception of the R976H mutant, which has a delayed release as indicated by the slightly longer half-rate of exchange t½ = 9.5 s (Fig. 3A and Table 2). This increased t½ may be due to its reduced affinity for binding G/T mismatches.
FIGURE 3.
ADP to ATP exchange and dissociation from a mismatch. A, exchange of ADP for ATP. hMSH2-hMSH6 protein was pre-bound to [3H]ADP. Exchange of ADP for ATP upon the addition of a 41-bp mismatch (G/T) containing oligonucleotide and excess ATP was determined by filter binding. Data points represent the average of three trials and were fit to exponential decay. B, ATP-dependent dissociation from a mismatch. 50 nm concentration of each protein was pre-bound to a 41-bp mismatch (G/T) containing oligonucleotide in the absence of ATP. The dissociation curves for wild-type and hMSH6 mutant heterodimers in the presence of 250 μm ATP are displayed as a function of the percentage of protein initially bound to the mismatch.
The mismatch-induced ADP → ATP exchange also results in the release of MSH2-MSH6 from the mismatch in the form of a hydrolysis-independent sliding clamp that dissociates from the DNA via a free end (9, 10). SPR was used to assess ATP-dependent dissociation from a mismatch. Wild-type and all missense mutant proteins quickly dissociate from the DNA in the presence of ATP with nearly indistinguishable off-rates (Fig. 3B). That the G566R and D803G mutants are able to dissociate in the presence of ATP despite their reduced affinity for ATP indicates that their ATP binding ability at high concentrations of ATP is sufficient to induce dissociation, although whether they undergo a conformational change to a sliding clamp cannot be determined by these experiments.
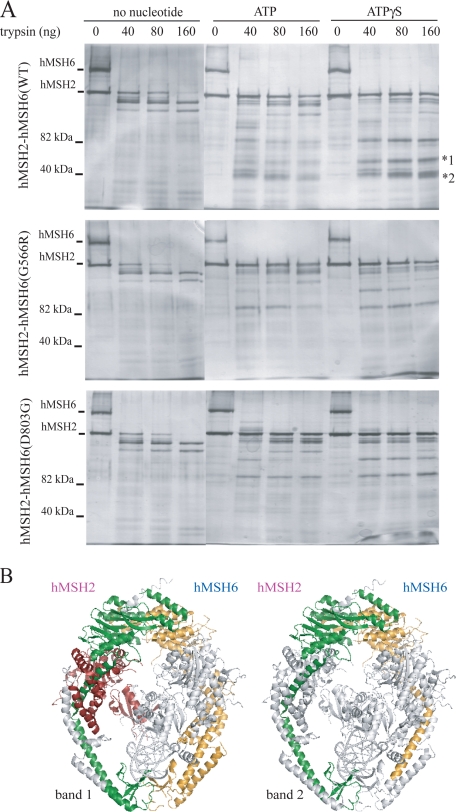
hMSH6 Missense Mutants Alter Nucleotide-induced Conformational Change in hMSH2-hMSH6—As the G566R, D803G, and V878A mutations appear to uncouple mismatch-stimulated hydrolysis from mismatch binding despite their locations far removed from the ATPase and mismatch binding sites (Fig. 1C), we began to investigate whether these mutations can affect ATP-dependent conformational changes. We performed partial proteolysis of the wild-type and mutant heterodimers with increasing concentrations of trypsin. The accessibility of trypsin digestion sites in wild-type hMSH2-hMSH6 is distinguishable upon the addition of ATP or ATPγS indicating a significant conformational change (Fig. 4A). Western blot analysis reveals that the majority of novel fragments in the ATP- and ATPγS-containing lanes are fragments of hMSH6, whereas the majority of hMSH6 fragments in the absence of nucleotide run off an 8% acrylamide gel indicating that hMSH6 is likely largely unstructured in the absence of nucleotide (supplemental Fig. 4A). However, we cannot rule out that the polyclonal antibodies used for hMSH2 and hMSH6 fail to recognize all digested fragments. Upon addition of ATP and ATPγS to the G566R and D803G heterodimers, altered proteolytic patterns are observed compared with wild type (Fig. 4A). At least two prominent fragments between 35 and 55 kDa observed in the wild-type digestion are not present for the mutants suggesting a different conformational state. This altered conformation is observed for V878A and H1248D as well, although the S144I proteolysis pattern appears similar to wild-type protein (supplemental Fig. 4B).
FIGURE 4.
Partial proteolytic digestion of hMSH2-hMSH6. A, nucleotide-induced conformational changes in hMSH2-hMSH6 were observed by trypsin proteolysis. 1 μg of hMSH2-hMSH6 protein was incubated with 1 mm adenosine nucleotide where indicated and increasing amounts of trypsin. Wild-type and mutant heterodimers undergo conformational change in the presence of ATP and ATPγS, but mutant proteins (G655R and D803G) lack two major protected fragments (*1 and *2), suggesting an altered conformational state. B, peptide fragments identified by mass spectrometry analysis of bands 1 and 2 from wild-type partial proteolysis reactions mapped to the crystal structure of hMSH2-hMSH6 bound to a mismatch (25). The hMSH6 peptide fragment identified in each band is labeled in yellow. The hMSH2 peptide fragment identified in band 2 is labeled in green. Two predominant hMSH2 peptide fragments were identified in band 1 labeled in green and red. Note, the fragments are mapped to the crystal structure for illustrative purposes only. The experiments were performed in the absence of mismatched DNA and in the presence of adenosine nucleotide. Thus, the conformation of the affected domains shown in the figure does not necessarily reflect their conformation in the experiment.
To identify the peptides that are present in the wild-type fragments between 35 and 55 kDa, the two major bands were cut out from a gel and analyzed by mass spectrometry. Both bands consisted of peptides of hMSH2 and hMSH6 indicating that each band represented multiple protected peptides that co-migrated on the gel. The faster migrating protected species included an hMSH2 fragment and an hMSH6 fragment. They consisted of the entire ATPase domains as well as the long α-helices (labeled α-helix V in hMSH2 and α-helix DD in hMSH6 in Fig. 2 of Warren et al. (25)) that extend the length of the lever domain connecting the ATPase domain to the clamp domain (Fig. 4B). The slower migrating protected species include fragments that are made up of the same domains as above plus additional sequences in the clamp domain from hMSH2 and hMSH6. In addition, peptides from the N-terminal DNA binding and connector domains of hMSH2 were identified in this slower migrating band, likely representing a third protected fragment that co-migrates on the gel. The absence of these species in the mutant heterodimer reactions suggest that missense mutations in hMSH6 can alter the conformation of both hMSH6 and hMSH2 such that the ATPase and lever domains are more accessible to trypsin digestion.
DISCUSSION
Premature truncation and deletion mutations in the hMSH2 or hMSH6 gene result in loss of function of that gene product leading to cancer. The significance, however, of missense variants in these genes is less clear. Whether the variation contributes to disease phenotype and whether carriers of the variant are at increased risk are important clinical questions that may affect patient management. Any functional assessment of the variation can add valuable information regarding the likelihood of its contribution to pathogenicity (30). Functional screens such as complementation assays in yeast (18, 31) and human cells (26) have revealed some information about the function, or lack of function, of certain missense variants. Previously, we examined the biochemical properties of seven missense mutations of hMSH2, demonstrating that six of seven mutations significantly disrupted the biochemical function of the hMSH2-hMSH6 heterodimer (21). This study examines the effect of seven different single amino acid alterations in hMSH6 on heterodimer function. We have demonstrated that five of the seven alterations affect mismatch-stimulated ATPase activity, providing strong evidence that these variants are true disease-causing mutations, and carriers should be considered at-risk for disease development.
Examining the manner in which cancer-associated variants of MMR genes affect their molecular function may also help shed light on the details of the MMR mechanism. Although necessary for MMR function, the exact role of the mismatch stimulated ATPase of hMSH2-hMSH6 is still not fully understood (2). The heterodimer utilizes its adenosine nucleotide processing function to directly or indirectly associate the site of a DNA mismatch with that of a strand discrimination signal at a distant site. This strand discrimination signal results in the proper generation of an excision tract by which the mispaired base is removed. Three models have been proposed for how hMSH2-hMSH6 coordinates mismatch binding and adenosine nucleotide processing to perform this job. A static transactivation model proposes that the MutS homologs form a ternary complex with ATP and the MutL homologs at the site of the mismatch that is capable of interacting with a strand discrimination site in trans (32). Two other models involve movement of the MutS homologs away from the mismatch either via an ATP hydrolysis-dependent translocation (33) or through the formation of an ATP-dependent sliding clamp that diffuses along DNA in a hydrolysis-independent fashion (4, 10). The manner in which cancer-associated variants of hMSH6 affect the function of the hMSH2-hMSH6 heterodimer may unravel clues as to the mechanism by which these proteins connect the mismatch and strand discrimination sites.
Of the five mutants that affect mismatch-stimulated ATPase, G566R, D803G, V878A, R976H, and H1248D, only R976H and H1248D affect the affinity of the heterodimer to a G/T mismatch-containing substrate. This is in contrast to our previous study of seven hMSH2 missense variants in which the ATPase activity and mismatch affinity seemed closely linked (21). Thus, at least three of the hMSH6 mutations tested appear to uncouple mismatch recognition from ATPase stimulation.
The S144I and S285I mutations do not have a hydrolysis defect suggesting that adenosine nucleotide processing in these complexes is not disrupted despite these mutations being associated with cancer. The S144I mutation was identified in the germline of three nonrelated cancer families but in none of 400 control individuals tested (18, 19, 34) providing strong genetic evidence that this variant is significant for disease. Ser-144 lies on the DNA binding surface of a PWWP domain at the N terminus of hMSH6, but it does not affect binding of the N-terminal fragment to DNA (35). Interestingly, tumors from different patients carrying this mutation display microsatellite instability, high and low phenotypes, suggesting this mutation may affect MMR despite the lack of a biochemical defect in vitro. S144I may have a role in downstream protein-protein interactions or MMR regulation as Ser-144 has been detected as a phosphorylation site (36). The S285I mutation was identified in the germline of one cancer family but not in 199 control individuals. The wild-type allele was lost because of a deletion mutation in the tumor consistent with loss of hMSH6 function in the tumor (18).
The two mutants among the most deficient in ATPase efficiency, G566R and D803G, are affected in their affinity for ATP. A deficiency in ATP binding should result in a protein that is incapable of mismatch-dependent nucleotide exchange and should affect ATP-dependent sliding clamp formation (10, 37). However, both mutant heterodimers release ADP rapidly in the presence of a mismatch and excess ATP and dissociate from a mismatch-containing substrate in the presence of ATP. Thus, although the mutants bind to the mismatch stably in the absence of nucleotide, they are not stuck at the site of the mismatch in the presence of ATP despite their reduced affinity for ATP. Based on the disease phenotype of the patients carrying these germline mutations, this result may not be surprising. A mutant protein that can bind to a DNA mismatch, but not release in the presence of ATP, may be expected to behave in a dominant negative fashion (37). One might expect such a dominant negative mutant to exhibit different disease phenotypes such as an earlier age of onset or a different spectrum of tumors, including lymphomas and leukemias such as observed in rare cases of homozygous mutant patients for hMSH6 (38, 39). The G566R mutation was identified in the germline of a patient that was diagnosed with rectal cancer at the age of 62 (18). The D803G mutation was from a patient diagnosed with rectal cancer at the age of 64 (18).
What cannot be determined from these ATP dissociation experiments, however, is the manner in which the proteins dissociate from the mismatch DNA template. In the presence of ATP and magnesium, wild-type hMSH2-hMSH6 has been demonstrated to form a sliding clamp that freely diffuses along DNA in an ATP hydrolysis-independent manner and dissociates from a free end of the DNA template (9, 10). However, in the absence of magnesium, ATP can induce dissociation of hMSH2-hMSH6 from a mismatch directly in a manner that does not appear to involve sliding clamp formation (10). Future experiments will need to be performed with double-blocked end substrates to determine the manner of dissociation of the G566R and D803G mutants. These mutants may provide tools to further probe the mechanism of sliding clamp formation and downstream hMSH2-hMSH6 functions in MMR.
A number of reports suggest that a significant conformational change occurs in MSH2-MSH6 upon binding to DNA and nucleotides that is likely to be crucial for proper coordination of downstream events in MMR (6, 10, 25, 29). The hMSH2-hMSH6 crystal structure shows that the ATPase- and DNA-binding sites are at opposite ends of the heterodimer and joined together by the connector and lever domains (25). These intermediary domains likely coordinate the conformational changes that allow for communication between the ATP and mismatch binding domains. We have identified single amino acid substitutions in the connector (G566R) and lever (D803G and V878A) domains (Fig. 1C) of hMSH6 that disrupt proper coordination of nucleotide processing with DNA binding, suggesting that the allosteric signaling between the distant sites in the heterodimer is affected. Consistent with this hypothesis, these variants are not fully capable of undergoing the same ATP-dependent conformational changes that wild-type protein undergoes as measured by partial proteolysis. It would seem that these variants confer a less compact structural state upon nucleotide binding than wild-type protein, as protected peptide fragments, apparent when wild-type protein is incubated with ATPγS, are not present for the mutants. Mass spectrometry analysis of these altered peptide fragments indicate they include both the entire ATPase domains and long α-helices in the lever domain of hMSH2 and hMSH6 (Fig. 4B). These α-helices connect the ATPase domains with the clamp domains in both monomers and likely play a crucial role in coordinating signaling between the two domains. Further structural analysis of the G566R, D803G, and V878A mutants and surrounding amino acids may help us unravel the detailed mechanism by which the hMSH2-hMSH6 heterodimer communicates intramolecularly to direct MMR. The combined results from this study lead us to hypothesize that heterodimers containing these three mutants are incapable of undergoing the proper ATP-dependent conformational change on mismatched DNA necessary to interact with hMLH1-hPMS2 and carry out the downstream steps of MMR.
Supplementary Material
Acknowledgments
We thank Drs. Mikhail Levin, John Carson, and David Han for helpful discussions regarding this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant CA115783 (NCI). This work was also supported by the Wendy Will Case Cancer Foundation, Grant RSG-07-145-01-CCG from the American Cancer Society. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: MMR, mismatch repair; HNPCC, hereditary nonpolyposis colorectal cancer; ATPγS, adenosine-5′-O-(3-thiotriphosphate); SPR, surface plasmon resonance; h, human; WT, wild type; DTT, dithiothreitol.
References
- 1.Jiricny, J. (2006) Nat. Rev. Mol. Cell. Biol. 7 335-346 [DOI] [PubMed] [Google Scholar]
- 2.Li, G. (2008) Cell Res. 18 85-98 [DOI] [PubMed] [Google Scholar]
- 3.Haber, L., and Walker, G. (1991) EMBO J. 10 2707-2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gradia, S., Acharya, S., and Fishel, R. (1997) Cell 91 995-1005 [DOI] [PubMed] [Google Scholar]
- 5.Alani, E., Sokolsky, T., Studamire, B., Miret, J. J., and Lahue, R. S. (1997) Mol. Cell. Biol. 17 2436-2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackwell, L. J., Bjornson, K. P., and Modrich, P. (1998) J. Biol. Chem. 273 32049-32054 [DOI] [PubMed] [Google Scholar]
- 7.Blackwell, L. J., Bjornson, K. P., Allen, D. J., and Modrich, P. (2001) J. Biol. Chem. 276 34339-34347 [DOI] [PubMed] [Google Scholar]
- 8.Antony, E., and Hingorani, M. M. (2003) Biochemistry 42 7682-7693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendillo, M. L., Mazur, D. J., and Kolodner, R. D. (2005) J. Biol. Chem. 280 22245-22257 [DOI] [PubMed] [Google Scholar]
- 10.Gradia, S., Subramanian, D., Wilson, T., Acharya, S., Makhov, A., Griffith, J., and Fishel, R. (1999) Mol. Cell 3 255-261 [DOI] [PubMed] [Google Scholar]
- 11.Fishel, R. (1998) Genes Dev. 12 2096-2101 [DOI] [PubMed] [Google Scholar]
- 12.Duval, A., and Hamelin, R. (2002) Cancer Res. 62 2447-2454 [PubMed] [Google Scholar]
- 13.Lynch, H. T., Smyrk, T., and Lynch, J. (1997) Cancer Genet. Cytogenet. 93 84-99 [DOI] [PubMed] [Google Scholar]
- 14.Woods, M., Williams, P., Careen, A., Edwards, L., Bartlett, S., McLaughlin, J. R., and Younghusband, H. B. (2007) Hum. Mutat. 28 669-673 [DOI] [PubMed] [Google Scholar]
- 15.Vasen, H. F., Mecklin, J. P., Khan, P. M., and Lynch, H. T. (1991) Dis. Colon Rectum 34 424-425 [DOI] [PubMed] [Google Scholar]
- 16.Wijnen, J., de Leeuw, W., Vasen, H., van der Klift, H., Moller, P., Stormorken, A., Meijers-Heijboer, H., Lindhout, D., Menko, F., Vossen, S., Moslein, G., Tops, C., Brocker-Vriends, A., Wu, Y., Hofstra, R., Sijmons, R., Cornelisse, C., Morreau, H., and Fodde, R. (1999) Nat. Genet. 23 142-144 [DOI] [PubMed] [Google Scholar]
- 17.Hendriks, Y. M. C., Wagner, A., Morreau, H., Menko, F., Stormorken, A., Quehenberger, F., Sandkuijl, L., Moller, P., Genuardi, M., van Houwelingen, H., Tops, C., van Puijenbroek, M., Verkuijlen, P., Kenter, G., van Mil, A., Meijers-Heijboer, H., Tan, G. B., Breuning, M. H., Fodde, R., Winjen, J. T., Brocker-Vriends, A. H. J. T., and Vasen, H. (2004) Gastroenterology 127 17-25 [DOI] [PubMed] [Google Scholar]
- 18.Kolodner, R. D., Tytell, J. D., Schmeits, J. L., Kane, M. F., Gupta, R. D., Weger, J., Wahlberg, S., Fox, E. A., Peel, D., Ziogas, A., Garber, J. E., Syngal, S., Anton-Culver, H., and Li, F. P. (1999) Cancer Res. 59 5068-5074 [PubMed] [Google Scholar]
- 19.Berends, M. J., Wu, Y., Sijmons, R. H., Mensink, R. G., van der Sluis, T., Hordijk-Hos, J. M., de Vries, E. G., Hollema, H., Karrenbeld, A., Buys, C. H., van der Zee, A. G., Hofstra, R. M., and Kleibeuker, J. H. (2002) Am. J. Hum. Genet. 70 26-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plaschke, J., Kruger, S., Pistorius, S., Theissig, F., Saeger, H. D., and Schackert, H. K. (2002) Int. J. Cancer 97 643-648 [DOI] [PubMed] [Google Scholar]
- 21.Heinen, C. D., Wilson, T., Mazurek, A., Berardini, M., Butz, C., and Fishel, R. (2002) Cancer Cell 1 469-478 [DOI] [PubMed] [Google Scholar]
- 22.Abramoff, M., Magelhaes, P., and Ram, S. (2004) Biophotonics Int. 11 36-42 [Google Scholar]
- 23.Rezaul, K., Wu, L., Mayya, V., Hwang, S.-I., and Han, D. (2005) Mol. Cell. Proteomics 4 169-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eng, J., McCormack, A., and Yates, J. R., III (1994) J. Am. Soc. Mass Spectrom. 5 976-989 [DOI] [PubMed] [Google Scholar]
- 25.Warren, J. J., Pohlhaus, T. J., Changela, A., Iyer, R. R., Modrich, P. L., and Beese, L. S. (2007) Mol. Cell 26 579-592 [DOI] [PubMed] [Google Scholar]
- 26.Kariola, R., Raevaara, T. E., Lonnqvist, K. E., and Nystrom-Lahti, M. (2002) Hum. Mol. Genet. 11 1303-1310 [DOI] [PubMed] [Google Scholar]
- 27.Guerrette, S., Wilson, T., Gradia, S., and Fishel, R. (1998) Mol. Cell. Biol. 18 6616-6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martik, D., Baitinger, C., and Modrich, P. (2004) J. Biol. Chem. 279 28402-28410 [DOI] [PubMed] [Google Scholar]
- 29.Mazur, D. J., Mendillo, M. L., and Kolodner, R. (2006) Mol. Cell 22 39-49 [DOI] [PubMed] [Google Scholar]
- 30.Ou, J., Niessen, R., Lützen, A., Sijmons, R., Kleibeuker, J., de Wind, N., Rasmussen, L., and Hofstra, R. (2007) Hum. Mutat. 28 1047-1054 [DOI] [PubMed] [Google Scholar]
- 31.Drotschmann, K., Clark, A. B., and Kunkel, T. A. (1999) Curr. Biol. 9 907-910 [DOI] [PubMed] [Google Scholar]
- 32.Junop, M. S., Obmolova, G., Rausch, K., Hsieh, P., and Yang, W. (2001) Mol. Cell 7 1-12 [DOI] [PubMed] [Google Scholar]
- 33.Allen, D. J., Makhov, A., Grilley, M., Taylor, J., Thresher, R., Modrich, P., and Griffith, J. D. (1997) EMBO J. 16 4467-4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, Y., Berends, M. J., Mensink, R. G., Kempinga, C., Sijmons, R. H., van Der Zee, A. G., Hollema, H., Kleibeuker, J. H., Buys, C. H., and Hofstra, R. M. (1999) Am. J. Hum. Genet. 65 1291-1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laguri, C., Duband-Goulet, I., Friedrich, N., Axt, M., Belin, P., Callebaut, I., Gilquin, B., Zinn-Justin, S., and Couprie, J. (2008) Biochemistry 47 6199-6207 [DOI] [PubMed] [Google Scholar]
- 36.Olsen, J. V., Blagoev, B., Gnad, F., Macek, B., Kumar, C., Mortensen, P., and Mann, M. (2006) Cell 127 635-648 [DOI] [PubMed] [Google Scholar]
- 37.Hess, M. T., Mendillo, M. L., Mazur, D. J., and Kolodner, R. D. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 558-563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bandipalliam, P. (2005) Fam. Cancer 4 323-333 [DOI] [PubMed] [Google Scholar]
- 39.Poley, J.-W., Wagner, A., Hoogmans, M. M., Menko, F. H., Tops, C., Kros, J. M., Reddingius, R. E., Meijers-Heijboer, H., Kuipers, E. J., Dinjens, W. M. N., and Rotterdam Initiative on Gastrointestinal Hereditary Tumors (2007) Cancer 109 2349-2356 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.