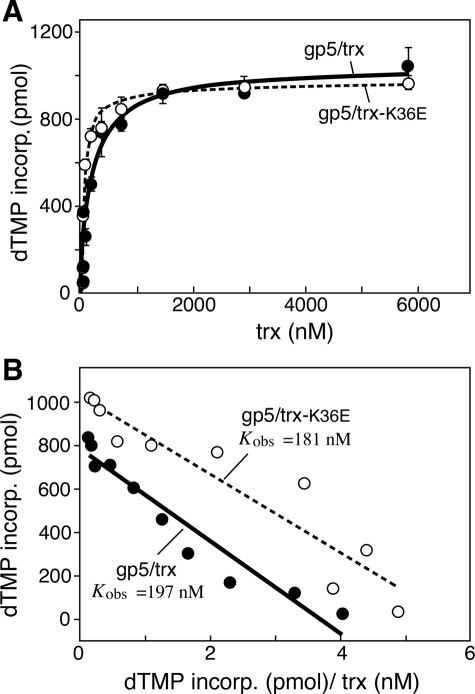
FIGURE 2.
Binding affinity of gp5 proteins to thioredoxins in the presence of M13 ssDNA. gp5-trx complexes were formed using A, gp5, with increasing amounts of trx (solid circles) or trx(K36E) (open circles). The polymerase activity of the complexes was measured as described under “Experimental Procedures.” DNA polymerase reactions (10 μl) contained 50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 5 mm DTT, 50 mm NaCl, 500 μm each of dATP, dCTP, dGTP, and [3H]dTTP (2 cpm/pmol), 50 μg/ml bovine serum albumin, 10 nm primed M13 ssDNA, and 10 nm gp5 or gp5-loop A or gp5-loop B or gp5-loop AB. Increasing amounts of trx or trx-K36E were added to each reaction as indicated, and the reactions were carried out at 37 °C. The amount of DNA synthesis for each reaction was determined by the amount of [3H]TMP incorporated over 3 min. B, data in A were used to generate Scatchard plots for binding affinity of the gp5-trx and gp5-trx-K36E complexes. The observed equilibrium constant (Kobs) for each complex was determined as the negative slope of the corresponding plot.