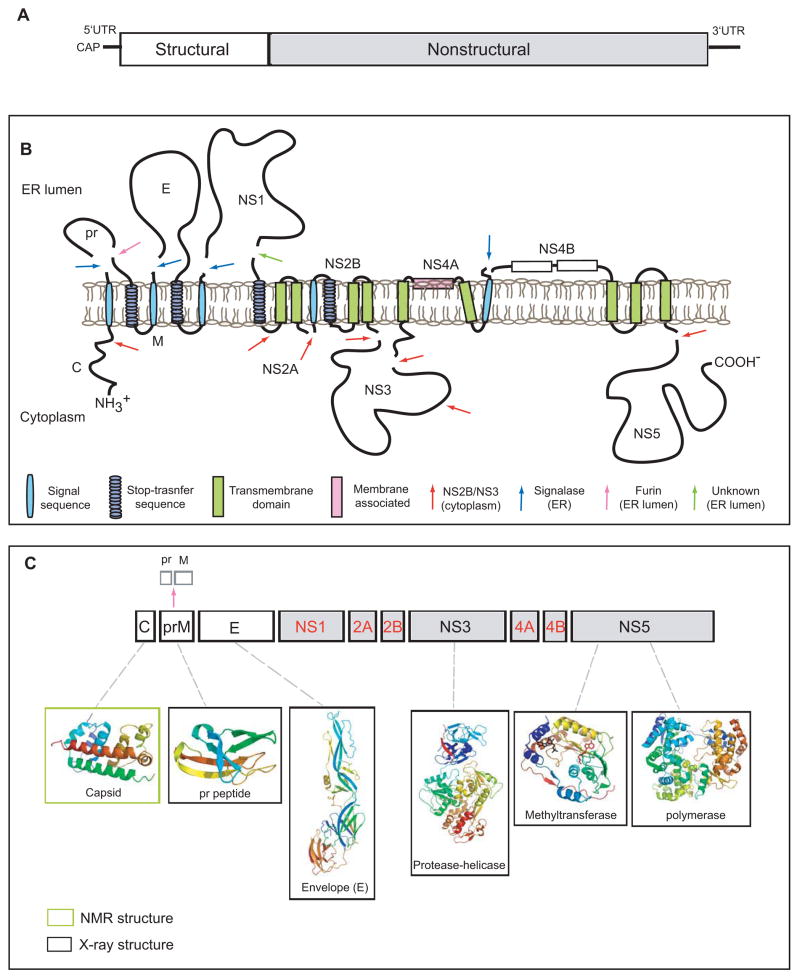
Figure 1. Schematic diagram of the dengue virus genome and polyprotein.
A. The viral genome is a positive sense RNA of ~11kb in length. It is capped at the 5′ end but lacks a poly-(A) at its 3′-end. The structural proteins (open boxes) are encoded at the 5′ one third of genome followed by the non-structural proteins (grey boxes). B. Membrane topology of the polyprotein. The viral RNA is translated as a polyprotein and processed by cellular and viral proteases (denoted by arrows). The structural proteins include capsid (C), membrane protein (prM/M) and envelope (E). prM and E are released from the polyprotein by signalase cleavage in the ER, but remain anchored on the luminal side of the membrane. The C is also anchored in the ER membrane (on the cytoplasmic side) by a conserved hydrophobic signal sequence at its C-terminal end. This signal sequence is cleaved by the viral NS2B-NS3 protease. During virus maturation, prM is further cleaved by furin in the TGN into the pr peptide and M protein. The non-structural proteins are processed mainly by the viral protease NS2B-NS3 in the cytoplasm with the exception of NS1, which is released from NS2A by a yet unidentified protease in the lumen of the ER. NS2A/2B and NS4A/4B are anchored in the ER as transmembrane proteins. The topology of NS4A and NS4B are predicted through biochemical and cellular analyses [62,67]. C. Structural proteome of dengue virus. NMR and X-ray structures are shown for C, prM, E, NS3 (full-length) and the NS5 methyltransferase and polymerase domains (PDB identifiers: 1R6R, 3C5X, 1OKE, 2VBC, 2P1D, 2J7U, respectively)[4, 5–7, 12, 13, 18, 32, 36]. Structures are currently not available for the proteins denoted in red.