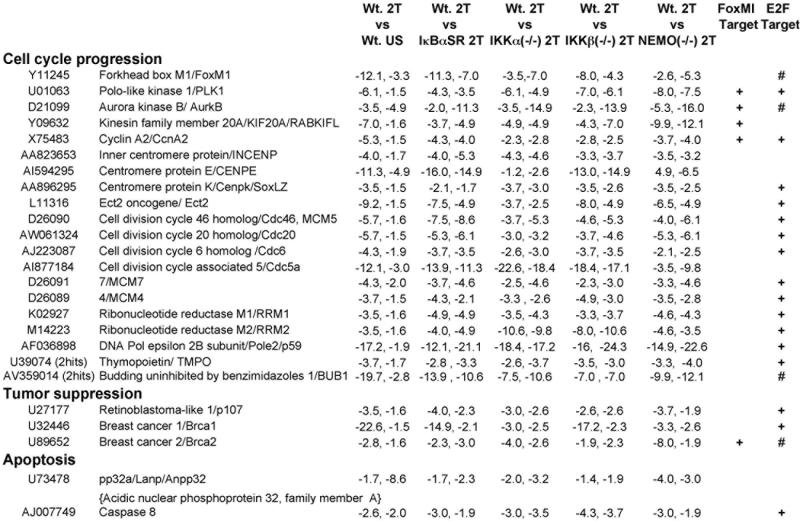
Figure 1. Genes repressed in response to TNFα with a dependency on NF-κΒ signaling.
Twenty-five genes, enriched in mediators of cell cycle progression, that were suppressed in response to TNFα stimulation dependent on canonical NF-κB signaling are shown. Cell lines, stimulation conditions and gene selection criteria are described in Materials and Methods and also in more detail in prior work (Li et al., 2002). Data for each gene is presented as fold change values from duplicate microarray screens with two independently derived stocks of immortalized MEFs. As elaborated in Materials and Methods, to simplify the presentation of this data negative integer fold change values for repressed genes were derived from their fractional fold change values,. Accession numbers and gene names are shown in columns one and two. TNFα dependency for gene repression is shown in Column 3 (Wt. MEF 2T vs. Wt. MEF US). Column 4 shows the NF-κB dependency of each repressed gene by comparing their expression in Wt. MEF 2T vs. Wt. MEF + IκBαSR 2T (i.e., Wt. MEF 2T : Wt. MEF + IκBαSR 2T). IKK signalsome subunit dependencies are shown in an analogous way in columns 5-7 displaying Wt. MEFs 2T MEFs vs. IKKα(−/−) 2T, vs. IKKβ(−/−) 2T and vs. NEMO(−/−) 2T MEFs respectively. Genes with 2 hits were identified by multiple Affymetrix oligo probes corresponding to distinct regions within the same gene with the data for one hit shown. In the two far right columns genes which have been reported to be direct targets of the FoxM1 and E2F transcription factors are indicated by + signs (Bindra and Glazer, 2006; Costa, 2005; Ishida et al., 2001; Wang et al., 2005; Yang et al., 2007), and those marked by a # sign were found to have E2F DNA binding sites in their promoters.