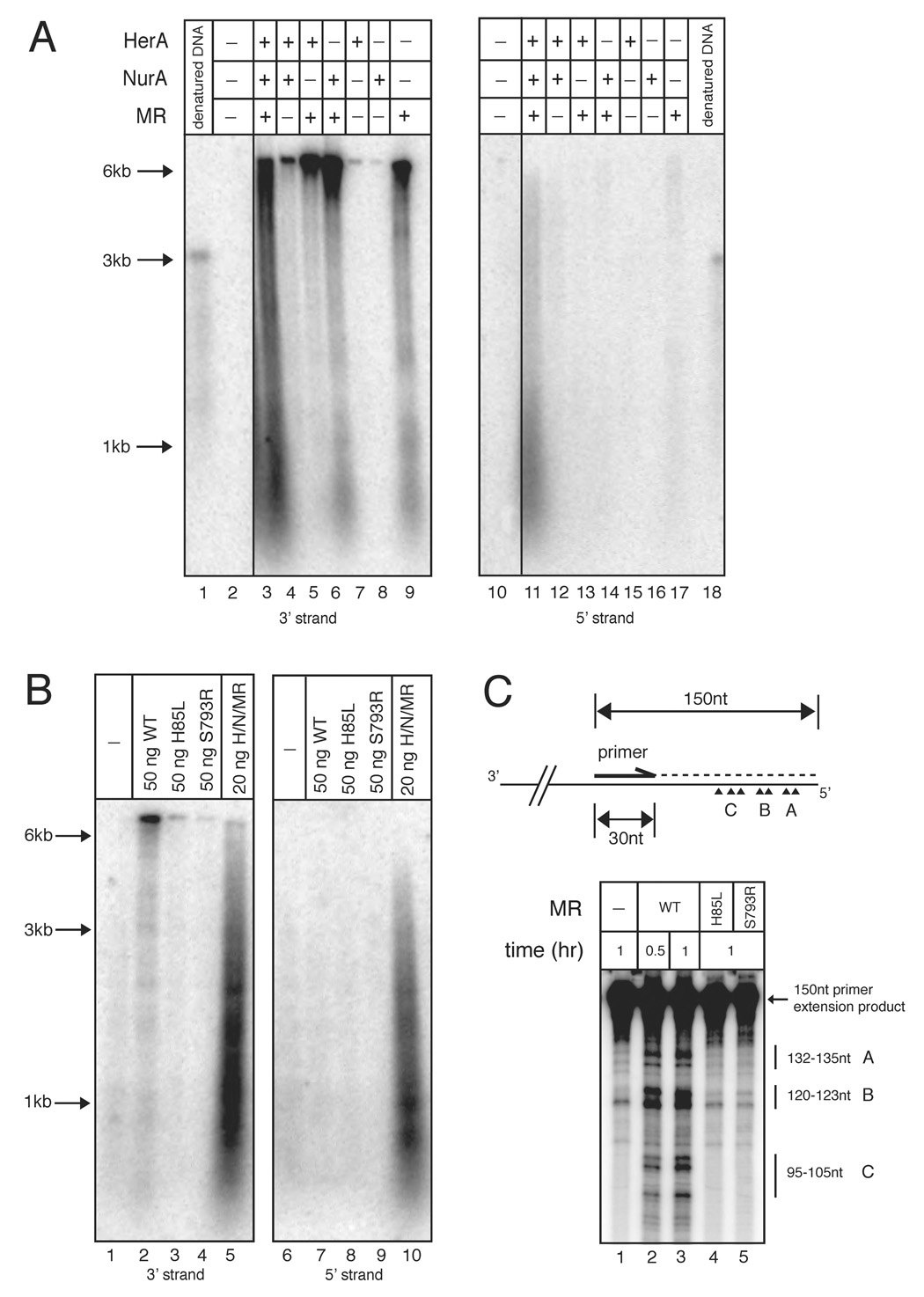
Figure 4. Mre11/Rad50 nuclease activity generates short 3’ overhangs.
(A) Nuclease assays were performed as in Fig. 2 but with 13.4 nM wild-type HerA, 96.2 nM wild-type NurA, and 16.5 nM wild-type MR as indicated. Reactions were incubated at 65°C for 1 hr.
(B) Reactions performed as in (A) but with 16.5 nM MR wild-type, M(H85L)R, or MR(S793R) proteins. Reactions in lanes 5 and 10 contained 5.4 nM HerA, 38.4 nM NurA, and 6.6 nM MR wild-type proteins. Reactions in lanes 1–4 and 6–9 were incubated at 65°C for 1 hr. Reactions in lanes 5 and 10 were incubated at 65°C for 15 min.
(C) (Top) Schematic of primer extension reaction. For simplicity, only the bottom strand of pTP163 is shown. The lengths of the full-length primer extension product and primer are indicated. The internally labeled primer extension product is indicated by the dashed line. Major MR resection sites are indicated as arrow heads and grouped into A, B, and C subgroups containing 2 or 3 cut sites. (Bottom) P. furiosus Mre11/Rad50 resection reactions were performed with 2.3 nM pTP163 linearized with SacI and 165 nM Mre11/Rad50 at 65°C for 30 min or 1 hr as indicated. Reactions were then used in primer extension reactions and the primer extension products were analyzed on 10% polyacrylamide denaturing sequencing gels. The position of the 150 nt primer extension product and the various resected products are indicated. Single-stranded DNA markers are indicated. For the full-size gel image, see Fig. S7.