Abstract
A cold-inducible transposon called Jordan has previously been used to tag and recover genes controlling key aspects of Volvox development, including the process called inversion. In a search for additional genes, we isolated 17 new inversionless mutants from cultures grown at 24° (the temperature that activates Jordan transposition). These mutants were stable at 32°, but generated revertants at 24°. DNA blots revealed that one mutant had a transposon unrelated to Jordan inserted in invA (“inversionless A”). This new transposon, which we named Idaten, has terminal inverted repeats (TIRs) beginning with CCCTA, and upon insertion it creates a 3-bp target-site duplication. It appears to belong to the CACTA superfamily of class II DNA transposons, which includes En/Spm. No significant open reading frames were in the Idaten sequence, but we retrieved another element with Idaten-type TIRs encoding a protein similar to the En/Spm transposase as a candidate for an Idaten-specific transposase. We found that in five of the new inversionless strains we could not find any Jordan insertions causing the phenotype to possess insertions of an Idaten family member in a single locus (invC). This clearly indicates that Idaten is a potentially powerful alternative to Jordan for tagging developmentally important genes in Volvox.
TRANSPOSONS can be powerful tools for tagging and cloning genes that play interesting biological roles, but whose molecular nature is unknown. During the 1980s numerous transposable elements from maize, snapdragons, fruit flies, and various other organisms began to be used to tag genes of interest within the species in which each of the transposons originated (reviewed by Gierl and Saedler 1992; Ryder and Russell 2003). Later it was shown that several such transposons could be used for insertional mutagenesis and gene tagging in species other than the ones in which they had originated. For example, the maize Ac/Ds elements have proven useful for tagging genes in Arabidopsis, rice, tobacco, and various other plants (reviewed in Ramachandran and Sundaresan 2001), and the Tol2 transposon of medaka has been used to tag genes in zebrafish and various other vertebrates (Kawakami 2005; Balciunas et al. 2006; Hamlet et al. 2006)
More recently, transposon tagging has been used to great advantage to identify and clone genes that control key aspects of development in Volvox carteri, a simple multicellular green alga that is increasingly popular as a developmental genetic and evolutionary model system (Kirk and Nishii 2001; Schmitt 2003; Kirk 2005). The appeal of V. carteri as a developmental model springs from the apparent simplicity of its programs for cellular differentiation and morphogenesis. Each V. carteri adult possesses only two cell types: 2000–4000 small, biflagellate somatic cells embedded in the surface of a transparent sphere of extracellular matrix and ∼16 large asexual reproductive cells, called gonidia, that lie beneath the somatic-cell layer (Starr 1970). All of the cells of both types that will be present in an adult are produced during subdivision of a mature gonidium by a series of embryonic cleavage divisions—some of which are visibly asymmetric and set apart large gonidial precursors from small somatic-cell precursors. At the end of cleavage each embryo is inside out with respect to the adult configuration, but it then turns itself right-side out in a dramatic morphogenetic process called inversion.
Analysis of the molecular underpinnings of important aspects of V. carteri development has been greatly facilitated in recent years by the availability of an endogenous transposon, Jordan, which has the extremely useful property of responding to temperature stress with an elevated rate of transposition (Miller et al. 1993). Cold-induced insertional mutagenesis with Jordan led directly to the cloning and characterization of three loci that play centrally important roles in V. carteri development: the glsA gene, which is required for the asymmetric divisions that set apart the germ and somatic lineages (Miller and Kirk 1999), the regA gene, which is required for terminal differentiation of somatic cells (Kirk et al. 1999), and the invA gene, which encodes a kinesin that drives inversion (Nishii et al. 2003). In the course of such studies, however, mutants were often encountered that had properties consistent with insertions of cold-inducible transposons, but that could not be associated with novel Jordan insertions (S. M. Miller, personal communication; I. Nishii, unpublished results). This led us to believe that V. carteri must possess at least one additional cold-inducible transposon. Finding and using such a transposon will enable us to obtain genes that still remain unknown by the Jordan transposon-tagging system.
Here we report the isolation and the characterization of just such an element: a newly discovered cold-inducible, cold-revertible transposable element from V. carteri that we have named Idaten after the name of a guardian god well known for his powers of running and jumping. We first encountered Idaten as a 9.7-kb insertion in the previously characterized invA locus while we were attempting to use Jordan insertions to tag and recover additional genes that are required for inversion of V. carteri embryos. We then demonstrated that another, previously unknown inv locus could readily and repeatedly be tagged with a transposon in the Idaten family, thereby establishing that Idaten provides a second cold-induced transposon-tagging system that may be even more useful than the Jordan system for tagging and recovering genes of developmental importance.
MATERIALS AND METHODS
V. carteri strains and cultivation conditions:
The starting strain for the present studies was CRH22, a morphologically wild-type descendant of HK10 (the standard V. carteri female; Starr 1970) that has been used repeatedly in the past as the source of Jordan-tagged mutants (Kirk et al. 1999; Miller and Kirk 1999; Nishii et al. 2003). Cultures of CRH22 and its descendants were maintained in standard Volvox medium (SVM) in bubbler flasks at 32° (except where stated otherwise) on a 16-hr-light/8-hr-dark illumination cycle (Kirk and Kirk 1983). A combination of warm light and daylight fluorescent lamps were used to give a light intensity at the flask surface of 20,000–27,000 lm/m2 (∼35 W/m2).
Isolation of revertible inversionless mutants:
Three hundred spheroids of CRH22 were inoculated into a flask containing ∼300 ml of SVM and were cultivated for 12 days under the standard light/dark cycle, but at 24°, which is near the lower limit temperature for growth of V. carteri and which has been shown to stimulate transposition of the Jordan transposon (Miller et al. 1993; Miller and Kirk 1999). Inversionless mutants were isolated from phototactically separated samples, as previously described (Nishii et al. 2003). In brief, the organisms were harvested and placed at one end of a long, SVM-filled glass tube that was illuminated at the opposite end with a small fluorescent lamp. Several hours later, when most of organisms had moved to the illuminated end, they were removed and discarded, whereas the few that had remained at the dark end—whether because of phototactic, motile, or morphological defects—were collected and cultured at 32° for 2 days and then subjected to another round of phototactic separation. From the phototaxis-negative organisms left at the dark end of the tube, we isolated individuals with apparent inversion defects under a dissection microscope. Each such individual was then cultured in 1 well of a 24-well plate at 32°, to assess the genetic stability of its inversionless phenotype. Strains with stable, heritable defects at 32° were then tested for their ability to generate wild-type revertants at 24°. Strains with elevated reversion rates at 24° were identified as candidate transposon-tagged mutants. In total, ∼2500 organisms from 14 phototactically enriched cultures were screened, yielding 448 strains with heritable morphological defects. Then, 42 “Inv” strains with inversion defects that were stable in both morphology and heritability at 32° were selected and cultivated at 24°. Finally, 17 strains were selected as exhibiting a significantly elevated reversion rate at 24°.
Southern blot analysis:
With one exception that is noted below, genomic DNAs were prepared by the cetyl trimethyl ammonium bromide (CTAB) method previously described (Miller et al. 1993; Miller and Kirk 1999). DNA samples were digested for 2–8 hr at 37° with KpnI or XhoI (TOYOBO, Osaka, Japan) and electrophoresed in a 0.8% SeaKem GTG agarose gel (Cambrex Bio Science, Rockland, ME) with TAE buffer. Gels were stained with ethidium bromide, photographed, and then incubated for 10 min in 0.125 m HCl. The DNA was transferred onto a positively charged nylon membrane (Hybond-N+, GE Healthcare, Bucks, UK) in alkali transfer buffer (0.5 n NaOH, 0.6 m NaCl) using a vacuum blotter (model 785; Bio-Rad Laboratories, Hercules, CA). The membrane was washed in 2× SSC and cross-linked using HL-2000 Hybrilinker UV Crosslinker (UVP, Upland, CA).
Labeled probes were prepared by PCR amplification of subcloned gene fragments using the following primer sets and templates. Primers p10 (5′-GCA GGG ACG GTT CTG GAC T) and p18 (5′-AAT AAA AGT AAA CGA TAC CTC CTG T) were used with a full-length invA genomic clone as template, to amplify a 1240-bp invA-hybridizing fragment called “probe A.” Primers IF01 (5′-GTT GTC AAC GTG GCA TAA CAG CCA) and IR01 (5′-AGA GCC TAC TTG GCA GAT TCA GCA) were used with a full-length cloned Idaten as template, to amplify a 1107-bp Idaten fragment called “probe I.” Primers CF04 (5′-TAT GTA CAA CCT GCA GCG ACC ACA) and CR04 (5′-AGA CTA ACT GCC TTA CCG GCG TTT) were used with a gel-purified inverse PCR product obtained from strain InvC1 (as described below) as template, to amplify a 1062-bp invC fragment called “probe C.” All custom primers were obtained from Invitrogen Japan (Tokyo). Probe labeling, hybridization, and signal detection were performed with Gene Images Random Prime Labeling Module and CDP-Star Detection Module following the supplier's instructions (GE Healthcare, UK). Signals were visualized with the VersaDoc Imaging system model 5000 with Quantity One software (Bio-Rad) and Photoshop CS3 (Adobe Systems, San Jose, CA) was used for adjusting level and contrast of 16-bit raw images to be seen as 8-bit gray images on display and print.
Isolation of Idaten and Idaten-2 genomic clones:
Both Idaten (inserted in invA) and Idaten-2 (inserted in invC) were amplified by long PCR, using LA Taq with GC buffer I (TaKaRa, Shiga, Japan) under the following cycling conditions: 1 min at 94° (1×) and then 10 sec at 98° and 15 min at 68° (30×). The primer pair 363 (5′-TGT TTG CTG TGT AGG CCT TGC TTG AGG) and 368 (5′-GCG TAG TCT TCA CGG TGG TAG TGT ACT) was used for amplification of Idaten, and a different primer pair, Cf06 (5′-TCC TTG TCC CAG CAC GGA GT) and CF04 (5′-TAT GTA CAA CCT GCA GCG ACC ACA), was used for amplification of Idaten-2. The resulting PCR products were cloned into the TOPO-XL vector (Invitrogen, Carlsbad, CA).
DNA sequencing:
DNA was sequenced using the ABI 3730 DNA Analyzer, primarily with BigDye v3.1 (Applied Biosystems, Foster City, CA), using the M13 forward, M13 reverse, and primers used for PCR, and extension by primer walking. For templates that were difficult to sequence because they were GC rich, or tended to form hairpin loops, alkaline denaturation (Hattori and Sakaki 1986) was performed before cycle sequencing with dGTP BigDye v3.0. Approximate lengths of the repetitive region III were estimated from the apparent size of the PCR amplification product on an agarose gel.
Genomic PCR:
Genomic DNAs were prepared from five InvA4 revertants either by the CTAB method referenced above or, in a few cases, by a whole-organism DNA prep method (Hallmann et al. 1997) that was adapted as follows: Twenty organisms were selected under the dissection microscope and transferred into 10 μl of lysis buffer (0.1 m NaOH, 2.0 m NaCl, 0.5% SDS). After 5 min at 95°, 200 μl of 50 mm Tris-HCl (pH 6.8) was added; and then 5 μl of the resulting lysate was used for PCR in a total volume of 50 μl. Genomic DNAs of these five revertants were amplified by PCR using primer pairs 363 (5′-TGT TTG CTG TGT AGG CCT TGC TTG AGG) and 368 (5′-GCG TAG TCT TCA CGG TGG TAG TGT ACT). The resulting fragments were purified with QIAquick PCR purification kit (QIAGEN, Tokyo) and sequenced directly, using the same primers.
Isolation of the region of the invC locus flanking the Idaten insertion:
The region of the invC gene flanking Idaten in the InvC mutant was isolated as follows: A 5.3-kb Idaten-containing KpnI restriction fragment that was present in InvC, but was missing from both CRH22 and all revertants analyzed, was gel purified with a QIAEXII gel extraction kit (QIAGEN). After self-ligation of 9.5 ng of the extracted DNA with T4 ligase (TOYOBO) at 37° for 1 hr in a total volume of 450 μl, 5 μl of the ligation mixture were used as a template for inverse PCR in a total volume of 50 μl. The reaction was heated to 94° for 5 min and then exposed to 25 cycles of 94° for 15 sec, 55° for 30 sec, and 68° for 3 min and then finally incubated at 68° for 6 min. The primers used were IF02 (5′-TGC TGA ATC TGC CAA GTA GGC TCT) and IR02 (5′-TGG CTG TTA TGC CAC GTT GAC AAC). Then 0.5 μl of the resulting product were used for nested PCR under the same conditions, using primers IF03 (5′-AAG CTC TAC GAC CGT GTG CTT CTT) and IR03 (5′-AGC TGA TAA TGA GCC CGT CTG ACA). An amplified fragment of the predicted size (∼4.2 kb) was purified from an agarose gel and sequenced directly using the IF03 and IR03 primers. The sequence of part of this amplified product matched part of a gene model in the V. carteri genome assembly (JGI protein no. 127353; location, Volca1/scaffold_1:2572794–2575397) that we subsequently named invC. DNA-blot analysis using a 1062-bp region of the cloned fragment as probe (probe C) confirmed that the isolated fragment represented the RFLP that was associated with the inversionless phenotype of the InvC mutant. This probe C was also used to determine whether any of the other new mutant strains contained insertions in invC. We retrieved and sequenced three EST clones corresponding to the invC gene from the JGI V. carteri database; these were CBGZ11879, CBGZ19301, and CBGZ25551. Their sequences were identical to the JGI gene model, except for the 3′-UTR. The Idaten insertion points in InvC strains were determined by PCR and direct sequencing using primer pairs of IR02 and primers designed within the invC locus as follows: Cr06 (5′-GCG TTA GGT AGC GCC TTG AAC AAT) for InvC1, Cf06 (5′-TCC TTG TCC CAG CAC GGA GT) for InvC2, InvC4, and InvC5, and CR04 (5′-AGA CTA ACT GCC TTA CCG GCG TTT) for InvC3.
Genome analysis:
To locate Idaten-related sequences within the V. carteri genome, 500-bp sequences from the left and right ends of Idaten were used separately to query the JGI V. carteri genome database (http://genome.jgi-psf.org/Volca1/Volca1.home.html) by BLASTN. The positions and directions of all hits with low E-value (<1 × 10−14; left end, 90 hits; right end, 58 hits) were analyzed. Sixteen Idaten-related pairs were found that were on the same scaffold, oriented in the same direction, and <50 kb apart (supplemental Table 1). The sequence between each pair was queried against the translated nucleotide database at the National Center for Biotechnology Information (NCBI, Bethesda, MD), using the TBLASTN algorithm to search for transposase-related genes. The CACTA, En/Spm transposase-like encoding sequence found between the Idaten ends located on scaffold 3 of the V. carteri genome assembly was aligned by MUSCLE on http://www.ebi.ac.uk/Tools/muscle/index.html (Edgar 2004) and shaded by BOXSHADE 3.21 (http://www.ch.embnet.org/software/BOX_form.html).
Microscopy:
Light-microscope images of adult organisms were acquired with a HR Plan Apo 1.6× objective on a SMZ1500 dissection microscope (Nikon, Tokyo) equipped with a ZEISS AxioCam MRc5 camera and Axiovision 4.6.3 software (Carl Zeiss, Oberkochen, Germany). DIC images of isolated embryos from synchronized culture of CRH22 and InvC were taken on a ZEISS Axio Imager Z1 microscope (EC Plan-Neofluar 40×/0.75 object) using the same camera and software. The areas of circle bounded by the outline of embryonic vesicle were measured using the NIH ImageJ analysis program (Abramoff et al. 2004).
RESULTS
Isolation of candidate transposon-tagged inversionless mutants:
The morphologically wild-type strain CRH22 (Figure 1A) was cultured at 24°, which activates Jordan transposition (Kirk et al. 1999; Miller and Kirk 1999). Then, after phototactic selection that enriched individuals either morphologically or physiologically unable to swim toward the light, we isolated individuals with morphological abnormalities under the dissection microscope (Nishii et al. 2003). Finally, we selected 17 strains that exhibited a significantly elevated reversion rate at 24°, which is a feature shared by many Jordan-induced mutants.
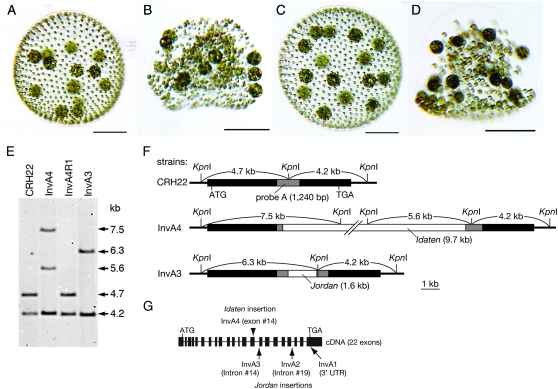
Figure 1.—
An Idaten transposon was trapped in the invA locus in mutant InvA4. (A–D) Young adults of four strains of V. carteri: (A) CRH22, the wild-type progenitor of all the mutants in this study; (B) strain InvA4; (C) strain InvA4R, a revertant derived from strain InvA4; and (D) strain InvA3, a mutant containing a Jordan insertion in the invA locus, as described by Nishii et al. (2003). Bars, 100 μm. (E) DNA blot of KpnI-digested genomic DNAs, hybridized to the invA probe (probe A) shown in F. Arrows and numbers indicate fragment sizes in kilobases. (F) Restriction maps of the invA loci present in the three strains indicated. Shaded rectangles, the portion of the invA gene that is represented in the hybridization probe that was used in E; solid rectangles, the rest of the invA gene; and open rectangles, transposon insertions. In CRH22 the 4.7-kb and 4.2-kb KpnI fragments cover the entire invA gene. In InvA4 the 4.7-kb fragment is interrupted by Idaten, which contains at least one KpnI site, and therefore the 4.7-kb band is replaced by two larger bands in E. In InvA3 the 4.7-kb fragment of invA is interrupted by Jordan, which lacks a KpnI site, and therefore the 4.7-kb band is replaced by a larger one in E. (G) The intron–exon map of the invA gene (length, 7608 bp; GenBank accession no. AB112467) with arrows indicating the sites of Jordan insertions in mutants InvA1, InvA2, and InvA3 described by Nishii et al. (2003). The arrowhead above invA indicates where Idaten was inserted in strain InvA4.
The invA locus trapped a novel transposable element:
Previously we had identified three inversionless mutants that had different Jordan insertions in the invA locus (Nishii et al. 2003). In a wild-type (CRH22) DNA digest on Southern blot, two fragments, 4.7 and 4.2 kb in length, that cover the entire invA locus are regularly observed (Figure 1, E and F) using an invA probe (probe A, Figure 1F). In strain InvA4, however, the 4.7-kb band was replaced by two bands, 5.6 and 7.5 kb in length (Figure 1E). This indicated that a DNA element at least 8.4 kb in length, and containing at least one KpnI site, had been inserted into the probe region of the 4.7-kb fragment (Figure 1F). This size, ≥8.4 kb, is much greater than the 1.6-kb Jordan insertions that have regularly been observed in the past (Kirk et al. 1999; Miller and Kirk 1999; Nishii et al. 2003), and it is also much larger than the largest member of the Jordan family that has been found in the V. carteri genome (3.6-kb Jordan-2; Miller et al. 1993).
In strain InvA4, inversion of the embryos is arrested shortly after it begins. As a result, InvA4 adults (Figure 1B)—just like InvA1 adults (Figure 1D)—are shaped rather like bowler hats, with high, rounded crowns and a curled, narrow brim. Five independent phenotypic revertants were recovered from InvA4 following cultivation at 24°; they all resembled wild-type V. carteri as adults (Figure 1C). Morphological reversion was accompanied by a DNA-level reversion (Figure 1E), suggesting essentially complete excision of the transposon. This indicates that the element inserted into the invA locus in strain InvA4 is a classical “cut-and-paste” type of transposable element.
Characterization of the novel transposon inserted in the invA locus in strain InvA4:
The restriction fragments present in the KpnI digest of InvA4 DNA (Figure 1E) indicated clearly that the unknown transposon had been inserted into the 4.7-kb KpnI fragment, somewhere within the short portion of that fragment that was complementary to the probe (Figure 1F). When primers designed to amplify that region of the gene were used in a PCR with either wild-type or revertant DNA as the template, the predicted ∼1.2-kb amplification product was obtained. In contrast, when InvA4 was used as the template, the product was >10 kb in length. When this large fragment was subcloned and sequenced (Figure 2A) it turned out to contain a 9.7-kb element inserted into the 14th exon of the invA gene (Figure 1G). The insert contained 36-bp terminal inverted repeats (TIRs) that begin with CCCTA, as do the Jordan TIRs (Figure 2, B and C), and three distinct repetitive regions (Figure 2A). We named this transposable element “Idaten” after a guardian god who is famous in Buddhist lore for his running and jumping ability.
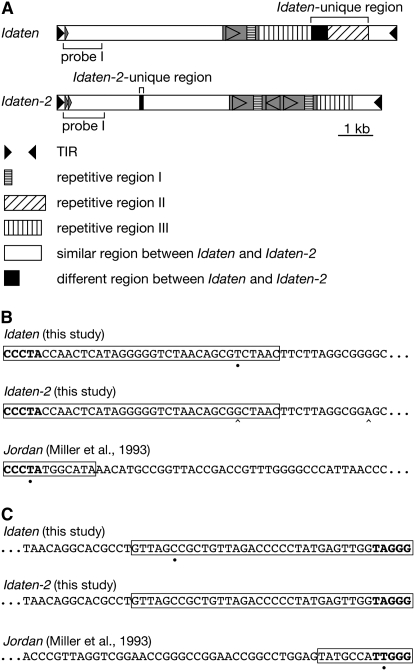
Figure 2.—
Structure of Idaten. (A) Schematic maps of Idaten and Idaten-2. The pair of solid triangles at opposite ends represent the terminal inverted repeats (TIRs). Both elements contain repetitive regions (striped boxes). Repetitive region I is enriched in C (52%) and T (31%) but there is no recurring simple motif. Repetitive region II has a 10-bp unit motif (5′-GGC AAG GGA G-3′ with slight variations). Repetitive region III has a 52-bp unit motif (5′-ATG GGT GCG ATG GTA AAC AGG CGG ATG TGT GGC ACG GCT ACC ATC GTA CCC A-3′ with slight variations). Repetitive region II and the adjacent 468-bp region in the right half of Idaten are Idaten-specific regions that are not shared by Idaten-2. Instead, Idaten-2 contains a 117-bp unique region in its left half. The shaded rectangle with internal shaded triangle indicates a region that is repeated as an inverted duplication only in Idaten-2. The region shown by a small rectangle located in the probe I region is duplicated in Idaten-2. The regions corresponding to probe I (the Idaten probe used in Southern blot analysis) are indicated at the bottom of the maps. (B and C) Sequences of the left end (B) and the right end (C) of Idaten, Idaten-2, and Jordan. The TIR sequences are boxed. The terminal five bases that are conserved between Idaten and Jordan are shown in boldface type. The left and right TIRs are fully complementary, except for individual bases of Idaten and Jordan that are indicated by dots on the bottom. Differences between Idaten and Idaten-2 are indicated by a ^ at the bottom.
Idaten is a TIR-containing, cut-and-paste transposable element that creates (as will be documented in a later section) a 3-bp target site duplication (TSD) upon insertion. TIR sequences and TSD size have been used for classification of the nine known transposon superfamilies in the TIR order (Wicker et al. 2007). Of those, the two superfamilies known to produce 3-bp TSDs are PIF-Harbinger (3 bp) and CACTA (2 or 3 bp). The PIF-Harbinger type differs from the CACTA type—and from Idaten—by having a target-site preference for TAA (Jurka and Kapitonov 2001). As the name suggest, transposons in the CACTA superfamily generally have this sequence at the beginning of their TIRs (Demarco et al. 2006), but the CCCTA sequence found at the beginning of the Idaten TIR (Figure 2, B and C) is also found in Jordan (Miller et al. 1993) and in a Tribolium castaneum transposon that belongs to the CACTA superfamily (Demarco et al. 2006). Together, its 3-bp TSD and its TIR motif indicate that Idaten—like Jordan and En/Spm—is a member of the CACTA superfamily of class II DNA transposons.
Idaten, like Jordan, appears to be a nonautonomous transposable element, because it contains no significant ORF. However, in a later section we will report a successful search for a candidate Idaten-specific transposase.
Multiple Idaten insertions identify a new gene, invC:
To assess the potential usefulness of Idaten as a tool for tagging novel genes of developmental importance, we performed DNA-blot analysis on various other inversionless mutants, using a 1107-bp sequence from one end of Idaten as a hybridization probe (probe I, Figure 2A, Figure 3E). We found that a mutant called InvC1 that never initiated inversion so it remained in the inside-out configuration as an adult (Figure 3A) produced a 5.3-kb KpnI fragment (Figure 3C; marked with red dots) that was not present in either the starting strain (CRH22) or three independent InvC1 revertants that were tested (Figure 3, B and C). A part of the InvC1-specific 5.3-kb DNA fragment was cloned by inverse PCR and sequenced. We found that it represented part of a gene present in scaffold 1 of the V. carteri genome sequence (http://genome.jgi-psf.org/Volca1/Volca1.home.html; protein no. 127353). We have named this gene invC. On a similar DNA blot hybridized with a 1,062-bp central portion of the invC sequence (probe C in Figure 3E), a >10-kb fragment was detected in DNA from the starting strain, CRH22, and from three independent InvC1 revertants (Figure 3D; marked with blue dots), whereas only the 5.3-kb fragment was detected in InvC1 DNA (Figure 3D; marked with red dots).
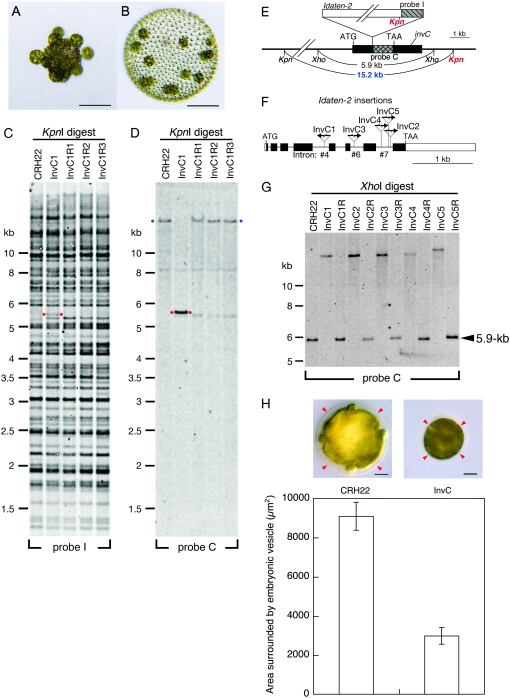
Figure 3.—
Transposon tagging with Idaten-2. (A) A young adult of the “fully inversionless” mutant, InvC1; note gonidia that are exposed on the outside. Bar, 100 μm. (B) A young adult of InvC1R, a revertant strain derived from InvC; note wild-type morphology. Bar, 100 μm. (C) A Southern blot of KpnI-digested DNAs from CRH22, InvC1, and three InvC1 revertants was hybridized to probe I, which was derived from Idaten as shown in E and Figure 2A. Red dots mark a 5.3-kb band that was detected in InvC1 but in none of the other strains. (D) A similar blot hybridized to probe C, which was derived from a piece of the genomic DNA adjacent to the Idaten insertion site, as shown in E. In addition to the original 5.3-kb fragment in InvC1 (red dots), probe C detected much larger fragments in CRH22 and all three revertants (blue dots). (Note that signals from the large bands are weak because transfer of such large DNA fragments from gel to membrane is generally incomplete.) (E) Restriction map of the invC region of the Idaten-2-tagged InvC1 mutant. The gray cross-hatched and hatched rectangles indicate the region of invC corresponding to probe C and the region of Idaten-2 corresponding to probe I, respectively. The black rectangles represent the rest of the invC locus and the white rectangle represents the rest of Idaten-2, which is shown at the top of its insertion site. The two red Kpn labels indicate the two KpnI sites that border the 5.3-kb fragment marked with red dots in C. The blue 15.2-kb label identifies the KpnI fragment that is seen in strains that lack Idaten-2 inserts in the invC locus (D). (F) Intron–exon map of invC. Rectangles indicate exons and connecting lines represent introns. Arrows show the Idaten-2 insertion sites in five InvC mutants. The direction of each arrow indicates the orientation of the Idaten-2 insert, relative to the orientation diagrammed in Figure 2A. (G) Southern blot analysis of XhoI-digested DNAs from five InvC strains. The 5.9-kb fragment that is detected by probe C in DNA samples from CRH22 and the five revertant strains is replaced by a much larger (>10-kb) fragment in the five InvC mutants, because of the Idaten-2 inserts in their invC genes. (H) Representative images of wild-type (CRH22; left) and InvC1 (right) embryos right before inversion show the size difference of the embryonic vesicle (arrowheads) that surrounds the embryo between two strains. It is noted that the InvC embryo appears to be tightly packed in the smaller vesicle, in which the space between the vesicle and the embryo is almost missing, while such space is obvious in wild type. In InvC, it is also hard to see individual gonidia and somatic cells compared to those of wild type. The bar chart indicates the morphometrical area surrounded by the vesicle with standard error bars (n = 30 for CRH22 and n = 16 for InvC1). Bar, 20 μm.
The entire wild-type invC locus is carried on a single 5.9-kb XhoI fragment (Figure 3, E and G, first lane), but because Idaten does not contain a XhoI site, in the InvC1 strain the XhoI fragment that hybridizes with probe C is ∼10-kb larger than the wild-type fragment (Figure 3G, second lane). To our surprise, when we tested the rest of our new inversion mutants, we found that four of them (which we then named InvC2, C3, C4, and C5) had XhoI fragments detected by probe C that were indistinguishable in size from the fragment in strain InvC1 (Figure 3G). The transposon-insertion sites in these four mutants were first estimated by DNA-blot analysis and then established precisely by PCR amplification and sequencing (Figure 3F). The fact that the transposon insertion points are all different in these five strains leaves no doubt that they are the result of five independent mutations.
The left-terminal sequences of these five invC insertions were found to be identical to one another, but all of them differed from the left-terminal sequence of the original Idaten element at a single nucleotide position (in Figure 2B). We named this new Idaten subtype Idaten-2.
The Idaten-2 element present in InvC2 was amplified by the long-PCR method, cloned, and sequenced. Although it is nearly identical in sequence to the original Idaten over much of its length, the two transposons differ in several ways (Figure 2A). For example, although they share two repetitive regions that are similar, Idaten contains a third repetitive region that has no counterpart in Idaten-2, and Idaten-2 has an ∼1-kb internal inverted duplication (shaded region in Figure 2A) not seen in Idaten.
The invC gene (Figure 3F) encodes a 401-residue polypeptide with 77% similarity to a gene model of Chlamydomonas reinhardtii (accession no. XP_001696547; Merchant et al. 2007) and both share a motif with members of the “LARGE” family of glycosyltransferase that are found in the mammalian Golgi apparatus (Grewal et al. 2005). This suggests that the InvC protein might be involved in constructing or modifying portions of the V. carteri glycoprotein-rich extracellular matrix, or possibly a membrane glycoprotein.
It is far from clear how a mutation in invC causes a total inhibition of inversion, but one observation we have made may be relevant: The glycoprotein-rich vesicle that surrounds all V. carteri embryos appears to be much smaller and tighter in the InvC mutant than it is in wild-type V. carteri (Figure 3H), and its compactness might conceivably interfere with inversion mechanically.
DNA sequences at sites of Idaten insertion and excision:
Sequencing of the transposon-insertion sites of the InvA4 mutant and four InvC mutants revealed 3-bp TSDs in all five of them (Figure 4, boldface type). Because nothing resembling a consensus sequence was found in these five TSDs (CAT, CGC, TAG, AGG, and TTT), it appears that Idaten and Idaten-2 have very weak insertion-site specificity—if any.
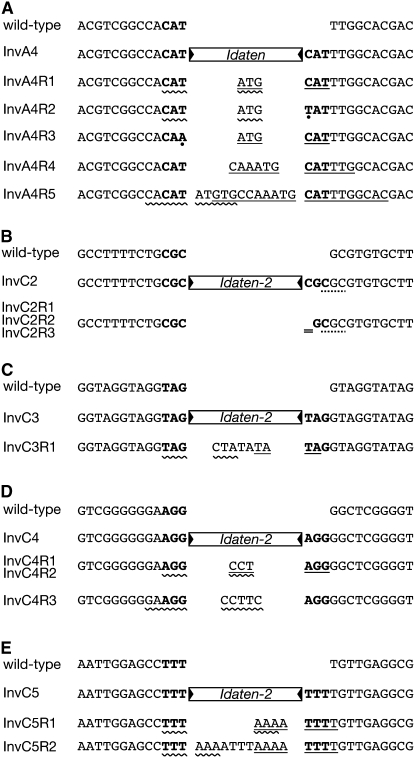
Figure 4.—
Modifications of target-site sequences that are associated with insertion and excision of Idaten and Idaten-2. The 3-bp TSDs are shown in boldface type. Open rectangles with closed triangles at their ends represent Idaten or Idaten-2 inserts. In the revertants, sequences in the middle are the footprints left following transposon excision. Portions of these footprints are antiparallel versions of the sequences to the left of the target site (wavy underlining) or the right side (straight underlining). (A) Mutant InvA4 and its revertants. Dots in two places indicate base changes in the TSD that were present following excision. (B) Mutant InvC2 and its revertants. All three revertants examined had a missing nucleotide in the right half of the TSD (double underline). The dotted line under three bases on the right side (CGC) calls attention to the fact that they are the same as the TSD, providing a possible reason why only this strain underwent a one-base deletion during excision. (C) InvC3 and its revertant. (D) InvC4 and its revertants. (E) InvC5 and its revertants. All revertants studied here were independent isolates.
Sequencing of the empty-donor sites in 15 revertant strains revealed that the sequence changes accompanying excisions were much more variable than those accompanying insertions (Figure 4). In most of the revertants we analyzed, excision of Idaten or Idaten-2 was accompanied by the appearance of a number of extra base pairs in the excision site. In all five InvA4 revertants that were examined, the number of additional base pairs was always a multiple of 3 (3, 6, or 12; Figure 4A); presumably this was because Idaten was inserted in one of the exons encoding the InvA protein (Figure 1G), and an excision event that left a footprint consisting of anything other than a multiple of 3 bp would have caused a frameshift that would almost certainly have precluded recovering the strain as a morphological revertant. There was no such constraint in the case of InvC insertions, however, because all of the InvC insertions were intronic. Consequently, the number of extra base pairs in the InvC footprints was highly variable: (−1), 3, 4, 5, 7, and 11.
In no case did any of the extra base pairs that were left as a footprint represent any part of the transposon that was excised. Instead, in every case some (or in most cases all) of these extra base pairs were antiparallel to (i.e., palindromic with) a number of base pairs flanking either one or both sides of the excision site (Figure 4; straight and wavy underlining). The only mutant strain in which extra base pairs were not inserted into the excision site was InvC2; in all three independent InvC2 revertants examined, no base pairs were added, but instead, in each revertant the same single base pair was lost at the right edge of excision site (Figure 4B; double underlining). (It is noteworthy that in this case the base pairs to the right of the TSD resembled the TSD itself; this might have somehow played a role in the loss of a base pair from the TSD during the excision). In addition to the extra base pairs that were inserted into the excision site in each of the InvA4 revertants, in two InvA4 revertants a base change occurred in the TSD during the excision (indicated by dots in Figure 4A; C → T for InvA4R2 and T → A for InvA4R3).
Finally, in one revertant not shown here (InvC5R3) a much more complicated set of changes occurred at the excision site: When Idaten-2 was excised from InvC5R3, so were 83 bp to its right, to be replaced by 25 bp taken from ∼1.1 kb downstream (plus one “gratuitous” A:T pair), while the 6 bp flanking the insertion site on the left side were duplicated in inverse order. Clearly, such a complicated set of sequence changes could only result in a morphological reversion when the transposon-insertion site was in an intron.
Search for candidate Idaten transposases in the V. carteri genome:
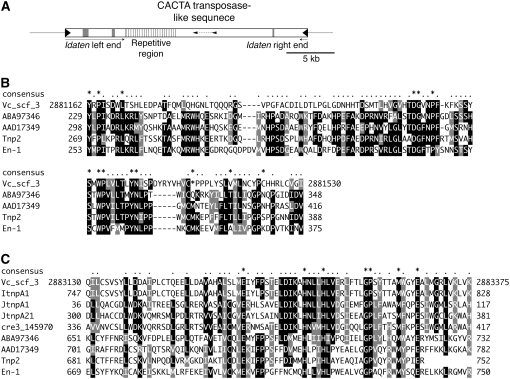
To determine whether any Idaten subtype in the genome might constitute a transposase-encoding autonomous transposon like maize En/Spm (Pereira et al. 1986), we used the opposite ends of the Idaten sequence separately to query the V. carteri genome with BLASTN. We found 16 Idaten-like elements ranging from 1.4 to 42 kb long on 12 scaffolds of the V. carteri genome (supplemental Table 1). Most of these elements contained long, unsequenced gap regions, reflecting the difficulty of sequencing the repetitive regions of Idaten and Idaten-2. But one of them, a 26-kb element on scaffold 3 (Figure 5A), contains two regions with homology (E-value < 10−2) to a number of genes that are annotated as putative CACTA, En/Spm-like transposases in the genomes of Oryza sativa and Arabidopsis thaliana and to a gene model (protein no. 145970; Merchant et al. 2007) with an En/Spm-like transposable element domain in the C. reinhardtii genome (Figure 5, B and C). Although this 2.2-kb region on scaffold 3 is the only element encoding part of a putative transposase that is flanked by both Idaten ends, nearly identical transposase-like sequences are also found in scaffolds 126 and 80 (supplemental Table 2), both of them downstream of an unpaired right-end sequence of Idaten. We named those regions of scaffolds 126 and 80 itnpA1 and itnpA2, respectively.
Figure 5.—
A candidate Idaten transposase found in the V. carteri genome. (A) Schematic structure of an Idaten-like element found in scaffold 3 of the V. carteri genome that contains two regions in the middle (small arrowheads) that show significant similarity to CACTA transposase-like genes. The ends (with solid triangles) are typical Idaten TIRs, as in Figure 2A. Shaded boxes indicate unsequenced gap regions. (B) An alignment of the upstream portion of the transposase-like element in A with related CACTA-transposase-like genes. Numbers at the ends of the Vc_scf_3 sequence are nucleotide numbers given to identify the region of scaffold 3 of the V. carteri genome sequence that encodes this deduced translation product. ABA97346, the GenBank accession no. of a putative transposon protein of the CACTA-En/Spm subclass from the O. sativa genome; length, 1051 aa. AAD17349, the GenBank accession no. of an A. thaliana element similar to A. majus TNP2; length, 817 aa. Tnp2 (GenBank accession no. CAA40555), A. majus transposable element homologous to the maize transposon En/Spm (Nacken et al. 1991); length, 752 aa. En-1 (GenBank accession no. AAA66266), Zea mays En/Spm-encoding transposase (Pereira et al. 1986); length, 897 aa. (C) Alignment of the downstream region of the CACTA transposase-like element in A with related transposase-like genes. The top sequence and the bottom four are identified in Figure 5B. cre3_14590, a hypothetical protein in the C. reinhardtii genome (accession no. EDP03893). The remaining three [ItnpA1 (JGI protein no. 100564; length, 966 aa), JtnpA1 (JGI protein no. 99812; length, 346 aa), and JtnpA21 (JGI protein no.108185; length, 618 aa)] are gene models developed in the V. carteri genome sequencing project (http://genome.jgi-psf.org/Volca1/Volca1.home.html). These gene models were predicted by the ab initio method, but because the upstream region depicted in B was not fully covered by their predicted cDNAs, they were not aligned for that region.
The preceding search was based on the assumption that the putative Idaten transposase should be associated with Idaten-end sequences. In an alternative approach, we simply searched the V. carteri genome for transposase-like sequences. In addition to the three described above, eight loci with low E-values (≤5 × 10−12) were found (supplemental Table 2), most of which had already been annotated as putative Jordan-transposition proteins. Although these loci are not flanked by Idaten ends, most of them are located near Idaten ends. In a third approach, we used the C. reinhardtii En/Spm-like gene (Figure 5C) as a query and 26 regions with low E-values (≤5 × 10−5) were identified (supplemental Table 3)—including the putative Idaten and Jordan transposition-protein-encoding genes, itnp and jtnp, respectively. It is interesting to note that in the C. reinhardtii genome only one other locus (one encoding JGI protein no. 179114) could be identified as having significant similarity (E-value ≤ 1) to this C. reinhardtii En/Spm-like sequence. This raises the intriguing possibility that the number of CACTA transposase-like sequences was expanded substantially during the evolution of V. carteri from a C. reinhardtii-like ancestor.
It is uncertain which itnp or jtnp gene—if any—encodes a full-length transposase responsible for Idaten transposition, because some of them appear to be pseudogenes that encode only a small part of a transposase, and other predicted gene models are difficult to align in their full length with any well-characterized transposases. Assuming that the evolution of transposases was rapid in the V. carteri lineage, a homology-based method of analysis may be of limited value. To determine which of these candidates may encode the actual function that transposes Idaten, more detailed molecular studies, such as cloning, expression analysis, and gene silencing will be required.
DISCUSSION
A new gene-tagging system for V. carteri:
The development of a transposon-tagging system based on the cold-inducible, cold-revertible transposon, Jordan (Miller et al. 1993), was a major turning point in V. carteri developmental biology because it provided the method that was used to clone three otherwise-elusive genes that play centrally important roles in V. carteri development, as described in the introduction. The present study began just as an attempt to exploit the Jordan-tagging system once again, to identify additional genes whose products are required for inversion of V. carteri embryos. With the aid of serendipity, what we discovered was something much better: the Idaten family of transposons, which provides a second cold-inducible, cold-revertible gene-tagging system that may be even more powerful than the system based on Jordan. This serendipitous discovery became possible only because one Idaten element happened to insert into a known gene, invA, in one cold-cultured V. carteri cell (Figure 1). Had that not happened, we probably never would have discovered the Idaten family.
Shared strengths and apparent differences of Jordan and Idaten:
The transcendent advantage of the Jordan and Idaten families of transposons for gene tagging is the fact that they are both cold inducible and cold revertible,but relatively stable at normal culture temperature. Consider the results reported here with respect to the invC locus: Cultivation of a wild-type strain of V. carteri at 24° yielded not just one, but five independent InvC mutants that failed to initiate embryo inversion (Figure 3F). That concurrence already made it seem virtually certain that a product of the invC gene must play an essential role in initiation of the inversion process. But then, when cultivation of those mutants at 24° generated not just one, but >10 independent strains in which the inversion defect and the transposon insertion had undergone coreversion, the importance of the invC product in the initiation of inversion was established beyond any reasonable doubt. In the absence of inducible reversion, establishing a causal relationship between the InvC genotype and phenotype would have been extremely difficult, if not totally impossible, because inversionless strains do not easily participate in sexual reproduction (Kirk 1998) and thus Mendelian cosegregation analysis would have been out of the question.
Although both the Jordan and Idaten transposon families exhibit cold-inducible transposition, the present study suggests that the Idaten family provides a much more efficient gene-tagging system than the Jordan family does. Of the 17 cold-revertible inversionless mutants that we isolated here, 13 turned out to be Idaten tagged, but no Jordan-tagged mutants were detected. Indeed, the Idaten-tagging system has been so efficient in our hands that we and other members of our research group have already cloned three additional, previously unknown inversion genes: invB, invD, and invE (N. Ueki, H. Toyooka, J. Kadota and I. Nishii, unpublished results).
One possible reason for the higher efficiency of the Idaten gene-tagging system might be caused by lower copy number for the transposon family. One would assume that the more members (active) of a given transposon family there are in the genome, the higher its insertional-mutagenesis rate might be. On the other hand, the more members there are (inactive as well as active members), the more hybridizing bands there will be on a DNA blot and thus the more difficult it will be to resolve a novel RFLP that is associated with a mutant phenotype. We count ∼50 Idaten-hybridizing bands on a typical DNA blot (Figure 3C). Using signal-intensity differences to correct for possible fragment overlap, we estimate that there may be ∼100 hybridizing fragments in the genome. This appears to be only about half as many bands as are present on a typical DNA blot probed with Jordan (Miller et al. 1993; Kirk et al. 1999; Miller and Kirk 1999; Nishii et al. 2003). Supporting the idea that novel bands may be easier to detect on a DNA blot probed with Idaten than on one probed with Jordan, we note that we were able to detect the InvC1-specific fragment not only on a blot of KpnI-digested DNAs (Figure 3C) but also on a blot of SphI-digested DNAs (data not shown). We are unaware of any cases in which a gene-specific Jordan fragment was similarly detected in two different kinds of DNA digests.
Although it is impossible to tell which of the fragments hybridizing to either Idaten or Jordan represent active vs. dead transposons, we know that at least two Idaten subtypes (Idaten and Idaten-2) are active and that both are detectable by the Idaten probe used in the present studies. In the future, however, the use of probes on the basis of type-specific Idaten and Idaten-2 sequences (Figure 2A) may allow us to detect mutant-specific RFLPs even more easily, because of the smaller number of hybridizing bands on the blots.
Even if Idaten is not routinely much better at tagging genes than Jordan is, having two kinds of potential gene-tagging agents at our disposal—rather than just one—should definitely be advantageous. In this regard, it is worth noting that 4 of the 17 inversionless mutants studied here had cold-revertible mutations that could not be attributed to either Idaten or Jordan insertions. This suggests that there may be at least one more family of cold-inducible transposons in V. carteri that remains to be discovered.
The cold-induction and transposition mechanisms of these transposons remain to be elucidated:
The molecular basis for the cold-inducibility of Jordan and Idaten is wholly unknown. It has been suggested that the low temperature-dependent transposition of the Antirrhinum majus (snapdragon) transposon, Tam3, involves methylation (Hashida et al. 2006), but we have no information whatsoever to indicate whether methylation changes might be involved in cold activation of transposition in Idaten or Jordan.
We also do not have any insight yet about the mechanistic details of insertion or excision of either family of transposons. However, in this regard it is important to note the similarities between the Idaten TSDs and excision footprints and those previously reported for Jordan (Miller et al. 1993). Both transposons generate TSDs that are direct repeats of 3 bp that are immediately adjacent to the insertion point. While the footprints left at the insertion by these two families of transposons when they excise are more variable than their TSDs, they exhibit very similar kinds of variations. Nine of the 11 Jordan revertants analyzed had a 3-bp footprint that was antiparallel to the TSD sequence, just as was observed here in five cases (InvA4R1, InvA4R2, InvA4R3, InvC4R1, and InvC4R2; Figure 4, A and D). The largest footprint seen in a Jordan revertant was 12 bp long, with one part derived from the sequence to the left of the insertion site and the rest derived from the sequence flanking the right side, and this same pattern was seen here in InvA4R5 (Figure 4A). Such similarities suggest that the mechanisms of insertion and excision of the two families of transposons must be rather similar, possibly because they are mediated by very similar transposase(s). In both cases, regions flanking the target site on both sides of the insertion site seem to be subject to being duplicated and/or recombined during both insertion and excision. Much additional work will be required to determine whether one of the candidate transposase genes we have partly identified here is actually involved in mediating Idaten transposition and if so, what the precise mechanisms of insertion and excision are.
Idaten and the future of V. carteri and its volvocine relatives as an evolutionary-genetic model system:
Interest in using V. carteri and its “volvocine” relatives as a model for exploring the evolution of multicellularity and cellular differentiation is as widespread as interest in using V. carteri as a developmental-genetic model (Kirk 1998, 2005; Michod 2007). The volvocine family tree includes several genera and many species that are intermediate in size and developmental complexity between unicellular C. reinhardtii and multicellular V. carteri. Although the tree is highly branched, there has been an overall tendency for organisms to become larger and more complex over the ∼50 million years since C. reinhardtii and V. carteri last shared a common ancestor (Rausch et al. 1989).
A major impediment to studying the evolution of developmental mechanisms in these algae is that we currently have extremely little genetic and molecular information about any of the intermediate organisms and that we lack methods for analyzing them at the genetic level. It is possible that the Idaten transposons could be used to circumvent that deficiency to some extent. In a best-case scenario, one would find taxa in which Idaten elements are present, can be activated by environmental stress, and can be used to begin tagging genes that are required in a developmental process—such as inversion—that is present in all volvocine taxa, but is visibly different in its details in different members of the group (Hallmann 2006). In a worst-case scenario, it might be found that none of the other volvocine taxa have an inducible Idaten system. In this case one would search for an autonomous Idaten element in V. carteri and attempt to use it for transgenesis and heterologous insertional mutagenesis, much as has been done with the maize Ac/Ds transposon in Arabidopsis (Long et al. 1993) or the Tol2 element of medaka in zebrafish (Kawakami et al. 2004).
Acknowledgments
We thank D. L. Kirk for insightful suggestions and critical reading of this manuscript. We thank all members of our research group for their assistance and helpful discussion, especially A. Nakazawa for preparation of genomic DNA from the revertants and maintaining many strains of our V. carteri and H. Toyooka for determining part of the Idaten-2 sequence. We also thank the Research Resource Center, RIKEN Brain Science Institute, for DNA sequencing. Sequencing of the V. carteri genome was performed by the Joint Genome Institute in Walnut Creek, CA (http://www.jgi.doe.gov/). This study was supported by the RIKEN Special Postdoctoral Researchers Program (N.U.) and the RIKEN Initiative Research Unit Program (I.N.).
References
- Abramoff, M. D., P. J. Magelhaes and S. J. Ram, 2004. Image processing with ImageJ. Biophotonics Int. 11 36–42. [Google Scholar]
- Balciunas, D., K. Wangensteen, A. Wilber, J. Bell, A. Geurts et al., 2006. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2 e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarco, R., T. M. Venancio and S. Verjovski-Almeida, 2006. SmTRC1, a novel Schistosoma mansoni DNA transposon, discloses new families of animal and fungi transposons belonging to the CACTA superfamily. BMC Evol. Biol. 6 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl, A., and H. Saedler, 1992. Plant-transposable elements and gene tagging. Plant Mol. Biol. 19 39–49. [DOI] [PubMed] [Google Scholar]
- Grewal, P. K., J. McLaughlan, C. Moore, C. Browning and J. Hewitt, 2005. Characterization of the LARGE family of putative glycosyltransferases associated with dystroglycanopathies. Glycobiology 15 912–923. [DOI] [PubMed] [Google Scholar]
- Hallmann, A., 2006. Morphogenesis in the family Volvocaceae: different tactics for turning an embryo right-side out. Protist 157 445–461. [DOI] [PubMed] [Google Scholar]
- Hallmann, A., A. Rappel and M. Sumper, 1997. Gene replacement by homologous recombination in the multicellular green alga Volvox carteri. Proc. Natl. Acad. Sci. USA 94 7469–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlet, M., D. Yergeau, E. Kuliyev, M. Takeda, M. Taira et al., 2006. Tol2 transposon-mediated transgenesis in Xenopus tropicalis. Genesis 44 438–445. [DOI] [PubMed] [Google Scholar]
- Hashida, S., T. Uchiyama, C. Martin, Y. Kishima, Y. Sano et al., 2006. The temperature-dependent change in methylation of the Antirrhinum transposon Tam3 is controlled by the activity of its transposase. Plant Cell 18 104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori, M., and Y. Sakaki, 1986. Dideoxy sequencing method using denatured plasmid templates. Anal. Biochem. 152 232–238. [DOI] [PubMed] [Google Scholar]
- Jurka, J., and V. Kapitonov, 2001. PIFs meet Tourists and Harbingers: a superfamily reunion. Proc. Natl. Acad. Sci. USA 98 12315–12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, K., 2005. Transposon tools and methods in zebrafish. Dev. Dyn. 234 244–254. [DOI] [PubMed] [Google Scholar]
- Kawakami, K., H. Takeda, N. Kawakami, M. Kobayashi, N. Matsuda et al., 2004. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell 7 133–144. [DOI] [PubMed] [Google Scholar]
- Kirk, D. L., 2005. A twelve-step program for evolving multicellularity and a division of labor. Bioessays 27 299–310. [DOI] [PubMed] [Google Scholar]
- Kirk, D. L., 1998. Volvox: Molecular Genetic Origins of Multicellularity and Cellular Differentiation. Cambridge University Press, New York.
- Kirk, D., and M. Kirk, 1983. Protein synthetic patterns during the asexual life cycle of Volvox carteri. Dev. Biol. 96 493–506. [DOI] [PubMed] [Google Scholar]
- Kirk, D., and I. Nishii, 2001. Volvox carteri as a model for studying the genetic and cytological control of morphogenesis. Dev. Growth Differ. 43 621–631. [DOI] [PubMed] [Google Scholar]
- Kirk, M., K. Stark, S. Miller, W. Muller, B. Taillon et al., 1999. regA, a Volvox gene that plays a central role in germ-soma differentiation, encodes a novel regulatory protein. Development 126 639–647. [DOI] [PubMed] [Google Scholar]
- Long, D., M. Martin, E. Sundberg, J. Swinburne, P. Puangsomlee et al., 1993. The maize transposable element system Ac/Ds as a mutagen in Arabidopsis: identification of an albino mutation induced by Ds insertion. Proc. Natl. Acad. Sci. USA 90 10370–10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant, S., S. Prochnik, O. Vallon, E. Harris, S. Karpowicz et al., 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michod, R. E., 2007. Evolution of individuality during the transition from unicellular to multicellular life. Proc. Natl. Acad. Sci. USA 104(Suppl 1): 8613–8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S., and D. Kirk, 1999. glsA, a Volvox gene required for asymmetric division and germ cell specification, encodes a chaperone-like protein. Development 126 649–658. [DOI] [PubMed] [Google Scholar]
- Miller, S. M., R. Schmitt and D. Kirk, 1993. Jordan, an active Volvox transposable element similar to higher plant transposons. Plant Cell 5 1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacken, W., R. Piotrowiak, H. Saedler and H. Sommer, 1991. The transposable element Tam1 from Antirrhinum majus shows structural homology to the maize transposon En/Spm and has no sequence specificity of insertion. Mol. Gen. Genet. 228 201–208. [DOI] [PubMed] [Google Scholar]
- Nishii, I., S. Ogihara and D. Kirk, 2003. A kinesin, invA, plays an essential role in Volvox morphogenesis. Cell 113 743–753. [DOI] [PubMed] [Google Scholar]
- Pereira, A., H. Cuypers, A. Gierl, Z. Schwarz-Sommer and H. Saedler, 1986. Molecular analysis of the En/Spm transposable element system of Zea mays. EMBO J. 5 835–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran, S., and V. Sundaresan, 2001. Transposons as tools for functional genomics. Plant Physiol. Biochem. 39 243–252. [Google Scholar]
- Rausch, H., N. Larsen and R. Schmitt, 1989. Phylogenetic relationships of the green alga Volvox carteri deduced from small-subunit ribosomal RNA comparisons. J. Mol. Evol. 29 255–265. [DOI] [PubMed] [Google Scholar]
- Ryder, E., and S. Russell, 2003. Transposable elements as tools for genomics and genetics in Drosophila. Brief. Funct. Genomic Proteomic 2 57–71. [DOI] [PubMed] [Google Scholar]
- Schmitt, R., 2003. Differentiation of germinal and somatic cells in Volvox carteri. Curr. Opin. Microbiol. 6 608–613. [DOI] [PubMed] [Google Scholar]
- Starr, R. C., 1970. Control of differentiation in Volvox. Symp. Soc. Dev. Biol. 29 59–100. [DOI] [PubMed] [Google Scholar]
- Wicker, T., F. Sabot, A. Hua-Van, J. Bennetzen, P. Capy et al., 2007. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8 973–982. [DOI] [PubMed] [Google Scholar]