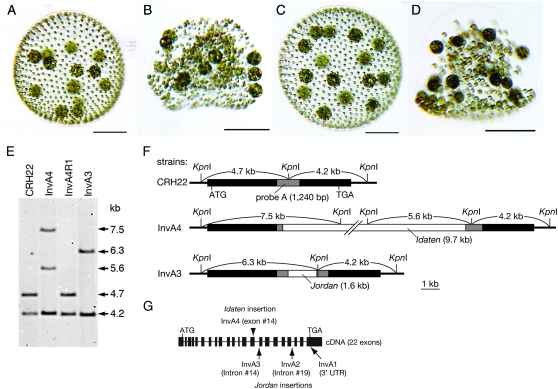
Figure 1.—
An Idaten transposon was trapped in the invA locus in mutant InvA4. (A–D) Young adults of four strains of V. carteri: (A) CRH22, the wild-type progenitor of all the mutants in this study; (B) strain InvA4; (C) strain InvA4R, a revertant derived from strain InvA4; and (D) strain InvA3, a mutant containing a Jordan insertion in the invA locus, as described by Nishii et al. (2003). Bars, 100 μm. (E) DNA blot of KpnI-digested genomic DNAs, hybridized to the invA probe (probe A) shown in F. Arrows and numbers indicate fragment sizes in kilobases. (F) Restriction maps of the invA loci present in the three strains indicated. Shaded rectangles, the portion of the invA gene that is represented in the hybridization probe that was used in E; solid rectangles, the rest of the invA gene; and open rectangles, transposon insertions. In CRH22 the 4.7-kb and 4.2-kb KpnI fragments cover the entire invA gene. In InvA4 the 4.7-kb fragment is interrupted by Idaten, which contains at least one KpnI site, and therefore the 4.7-kb band is replaced by two larger bands in E. In InvA3 the 4.7-kb fragment of invA is interrupted by Jordan, which lacks a KpnI site, and therefore the 4.7-kb band is replaced by a larger one in E. (G) The intron–exon map of the invA gene (length, 7608 bp; GenBank accession no. AB112467) with arrows indicating the sites of Jordan insertions in mutants InvA1, InvA2, and InvA3 described by Nishii et al. (2003). The arrowhead above invA indicates where Idaten was inserted in strain InvA4.