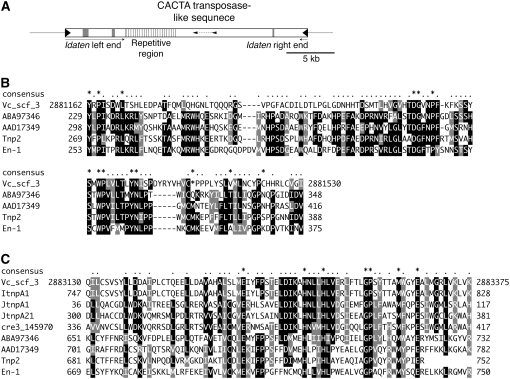
Figure 5.—
A candidate Idaten transposase found in the V. carteri genome. (A) Schematic structure of an Idaten-like element found in scaffold 3 of the V. carteri genome that contains two regions in the middle (small arrowheads) that show significant similarity to CACTA transposase-like genes. The ends (with solid triangles) are typical Idaten TIRs, as in Figure 2A. Shaded boxes indicate unsequenced gap regions. (B) An alignment of the upstream portion of the transposase-like element in A with related CACTA-transposase-like genes. Numbers at the ends of the Vc_scf_3 sequence are nucleotide numbers given to identify the region of scaffold 3 of the V. carteri genome sequence that encodes this deduced translation product. ABA97346, the GenBank accession no. of a putative transposon protein of the CACTA-En/Spm subclass from the O. sativa genome; length, 1051 aa. AAD17349, the GenBank accession no. of an A. thaliana element similar to A. majus TNP2; length, 817 aa. Tnp2 (GenBank accession no. CAA40555), A. majus transposable element homologous to the maize transposon En/Spm (Nacken et al. 1991); length, 752 aa. En-1 (GenBank accession no. AAA66266), Zea mays En/Spm-encoding transposase (Pereira et al. 1986); length, 897 aa. (C) Alignment of the downstream region of the CACTA transposase-like element in A with related transposase-like genes. The top sequence and the bottom four are identified in Figure 5B. cre3_14590, a hypothetical protein in the C. reinhardtii genome (accession no. EDP03893). The remaining three [ItnpA1 (JGI protein no. 100564; length, 966 aa), JtnpA1 (JGI protein no. 99812; length, 346 aa), and JtnpA21 (JGI protein no.108185; length, 618 aa)] are gene models developed in the V. carteri genome sequencing project (http://genome.jgi-psf.org/Volca1/Volca1.home.html). These gene models were predicted by the ab initio method, but because the upstream region depicted in B was not fully covered by their predicted cDNAs, they were not aligned for that region.