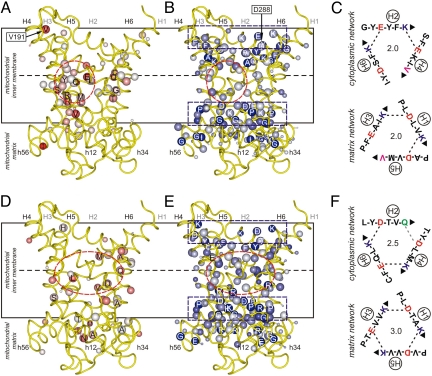
Fig. 2.
Asymmetry and symmetry in fungal phosphate transporters and mammalian uncoupling proteins. Average symmetry and conservation scores in the subfamily of (A and B) the phosphate transporters and (D and E) the uncoupling proteins projected onto a model of the bovine ADP/ATP transporter (8). The positive conservation score and average symmetry score are represented by the size and color of the Cβ atom, respectively (Fig. 1D). (A and D) Asymmetric residues with negative average symmetry scores. Residues in the cavity are labeled. (B and E) Symmetric residues with positive average symmetry scores. Highly symmetric residues are labeled. Dashed circles and rectangles indicate the location of the substrate binding site and networks, respectively. Green spheres represent residues that are absent from the repeat. Also shown are V191 and D288 (Fig. 1D). For all analyzed sequences the average of the standard deviations for all of the average symmetry scores of the cavity residues was 0.10. (C and F) The residues of the cytoplasmic (Upper) and matrix network (Lower) in ScPic2p and HsUCP1, respectively. The positively and negatively charged residues of the salt bridges are shown in blue and red, respectively. Deviating polar, aromatic and apolar residues are shown in green, orange and pink, respectively. The interaction energies of the network are quantified as the number of salt bridges, taking hydrogen bonds and cation-π interactions as half the interaction energy of a salt bridge, and van der Waals interactions as negligible.