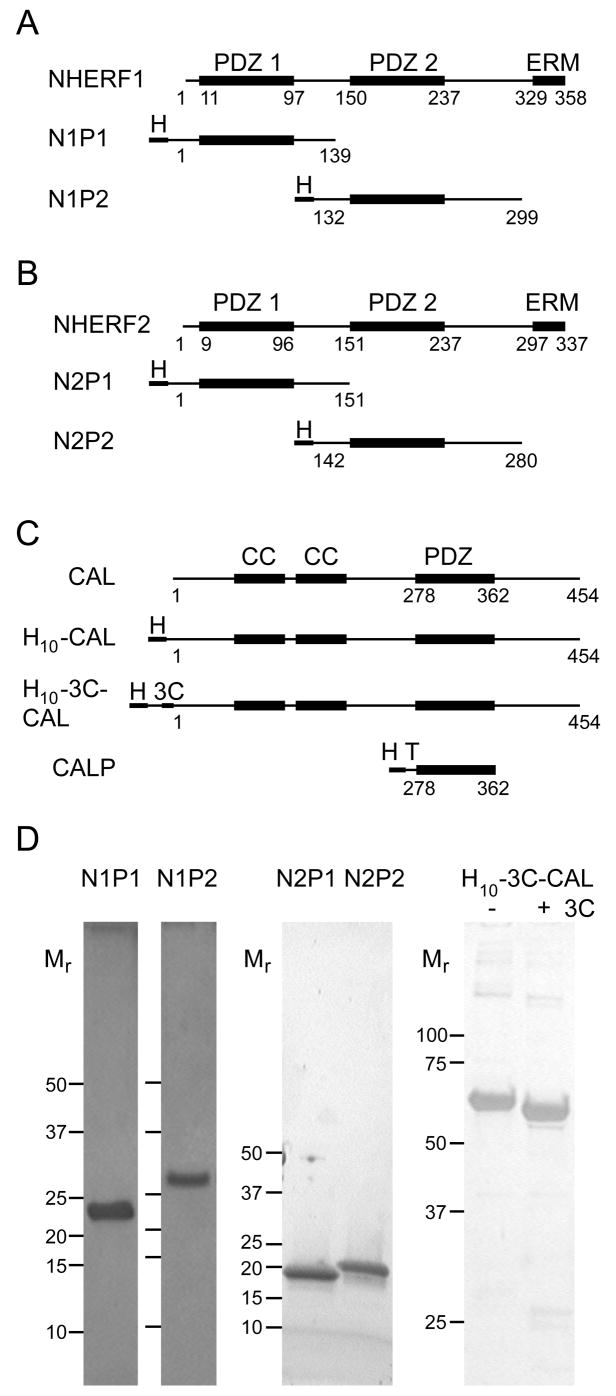
Figure 1. Purified proteins used in binding studies.
Schematic representations are shown for the domain structures (top line) of (A) NHERF1, (B) NHERF2, and (C) CAL, together with boundaries of the recombinant constructs used in this study (lower lines). PDZ, coiled coil (CC), and ezrin-radixin-moesin binding (ERM) domains are indicated above the native sequences (top line in A–C). The position of included domains (thick lines), decahistidine purification tags (H), TEV protease cleavage sites (T) and 3C-protease cleavage sites (3C) are indicated schematically for each construct. (D) Silver- (N1P1, N1P2, H10-3C-CAL) or Coomassie- (N2P1 and N2P2) stained SDS-PAGE gels are shown for previously unpublished protein constructs used in these studies. Mr standards are marked to the left of each lane. For the PDZ domains, Mr standards are shown at 50, 37, 25, 20, 15, 10 kD. For H10-3C-CAL, they are shown at 100, 75, 50, 37 and 25 kD. The dye front was run off the gel for H10-3C-CAL, to resolve more clearly the molar-mass shift before (−) and after (+) 3C-protease treatment. Protein Mr were separately validated by mass spectrometry for each construct.