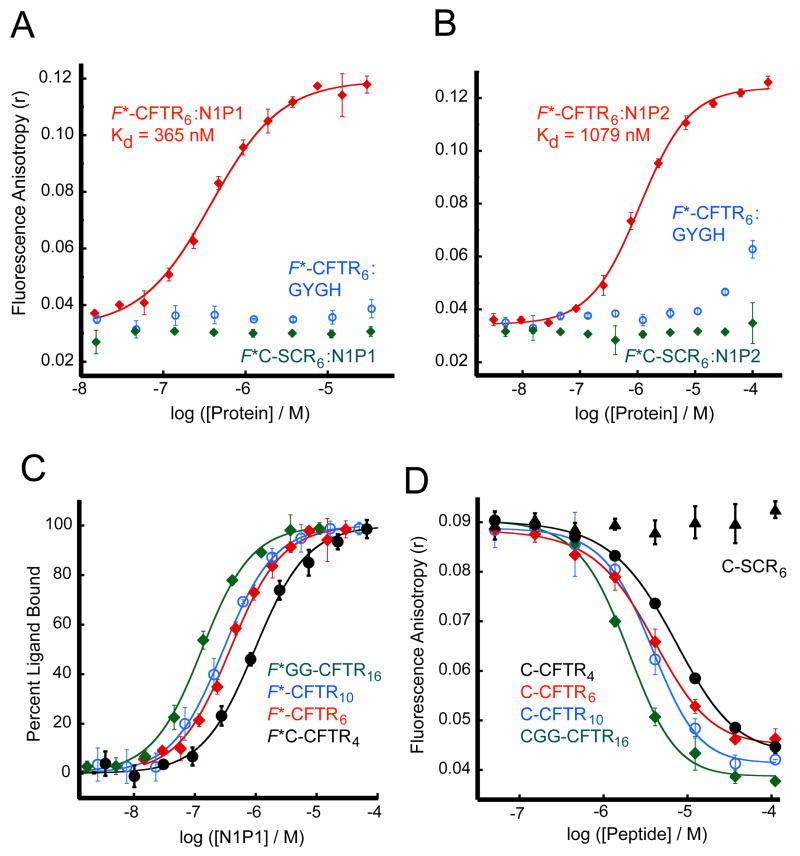
Figure 2. Solution-state binding assay reveals high affinity of the NHERF1 PDZ domains for the CFTR C-terminus.
(A and B) Fluorescence anisotropy data were obtained in the presence of increasing concentrations of NHERF1 PDZ1 (N1P1) (A) and PDZ2 (N1P2) (B) in the presence of reporter peptides corresponding to the C-terminal six residues of CFTR (F*-CFTR6, red) or a scrambled control sequence (F*C-SCR6, green), each covalently coupled to fluorescein. Curve fits (solid lines) yielded estimates of Kd of 365 ± 35 nM and 1079 ± 79 nM, respectively. Parallel titrations were performed with the GYGH mutants (blue) of the N1P1 (A) and N1P2 (B) domains. (C) Fluorescein-labeled peptides corresponding to the C-terminal four (F*C-CFTR4, black, filled circles), six (F*-CFTR6, red, filled diamonds), ten (F*-CFTR10, blue, open circles), or sixteen (F*GG-CFTR16, green, open diamonds) residues of CFTR were incubated at 30 nM each with increasing concentrations of the N1P1 domain, and the fluorescence anistropy determined. Curves were fit (solid lines) as in (A–B), yielding estimates of Kd shown in Table 2. (D) Residual fluorescence anistropy was determined for a mixture of F*-CFTR6 (30 nM) and N1P1 (1 μM ≈ 3 × Kd) incubated with increasing concentrations of unlabeled C-CFTR4 (black, filled circles), C-CFTR6 (red, filled diamonds), C-CFTR10 (blue, open circles), CGG-CFTR16 (green, open diamonds) or a scrambled control peptide (C-SCR6) (black, triangles), shown with logistic curve fits (solid lines). Values shown are mean ± SD, n=3.