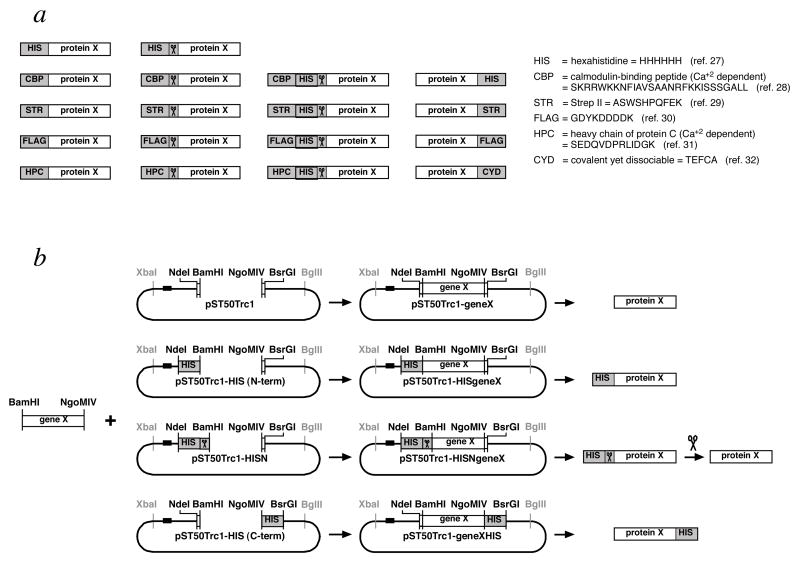
Figure 3.
Modular subcloning scheme facilitates incorporation of a wide range of affinity tag combinations. (a) Suite of affinity tags available for the pST50Trc1–4 transfer plasmids. 5 N-terminal, short affinity tags are available as single noncleavable (first column), single cleavable (second column), or double cleavable tags (third column). In addition, 4 C-terminal, noncleavable affinity tags are provided (fourth column). These 14 affinity tag combinations are available for each of pST50Trc1, pST50Trc2, pST50Trc3 and pST50Trc4 plasmids. The names and the sequences of the 6 different affinity tags are provided on the right. (b) Modular subcloning scheme for subcloning genes into pST50Trc1–4 plasmids. The same BamHI-NgoMIV coding region can be subcloned into an appropriate pST50Trc1–4 plasmid to express an untagged subunit (top row), an N-terminal, non-cleavable tagged subunit (second row), a N-terminal, cleavable tagged subunit (third row) or a C-terminal, non-cleavable tagged subunit (bottom row). Use of this subcloning scheme adds an N-terminal Gly-Ser (from the GGATCC BamHI site) and a C-terminal Ala-Gly (from the GCCGGC NgoMIV site) to the native protein. Other subcloning schemes are also possible including NdeI-BsrGI (for untagged subunits with no non-native amino acids), BamHI-BsrGI (for N-terminal tagged subunits only) and NdeI-NgoMIV (for C-terminal tagged subunits only). The scissors icon represents the tobacco etch virus (TEV) protease recognition site or the TEV protease itself. The horizontal black bar 5’ of the NdeI site in each plasmid represents the translational initiation signals such as the translational enhancer and Shine-Dalgarno sequence.