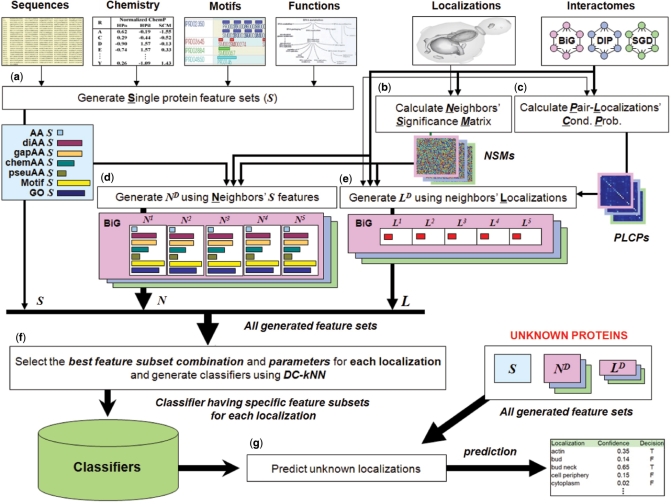
Figure 1.
Schematic overview of the integrated network-based framework. (a) Generation of single-protein feature vectors (Ss). Nine kinds of Si (AA, diAA, gapAA, three kinds of chemAA, pseuAA, Motif and GO) were generated for each protein Pi based on its sequence, chemical properties, motifs and functions. (b) Calculation of Neighbors’ Significance Matrixes (NSMs). These were calculated based on the number of distinct localizations covered by proteins falling along the path with the highest weight from a target protein to a neighbor protein (see Materials and methods section). (c) Calculation of PLCPs. They were calculated based on a weighted counting with normalization (see Materials and methods section). (d) Generation of network feature vector NiD. Each NiD was generated using up to D-th neighborhood's Ss with neighbors’ significance degrees from NSMs. (e) Generation of network feature vector LiD. Each LiD was generated using Pi's network neighbors up to distance D, weighted by NSMs and PLCPs to reflect each neighbor's significance and the conditional probabilities of interactions between localization pairs, respectively. (f) Model selection for each localization. The best combination of feature sets was selected for each localization based on a forward approach with the DC-kNN classifier. (g) Prediction of unknown localizations. After generating all feature vectors using all known localization and network information, a confidence degree and a decision (on whether an unknown protein has a specific localization or not) were computed for each localization.