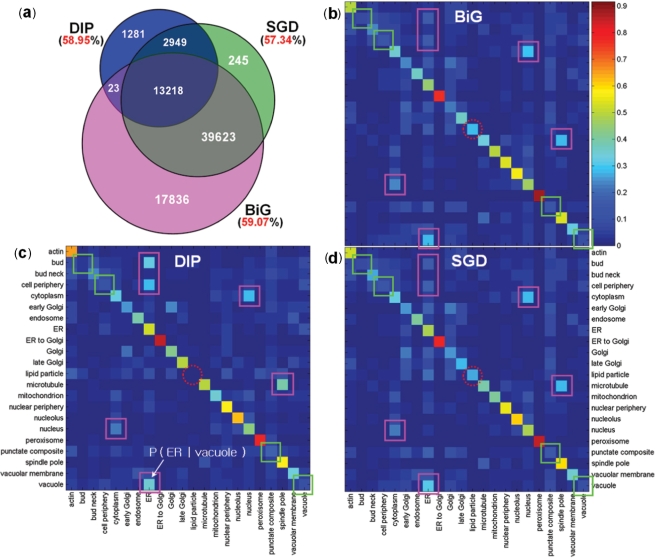
Figure 2.
Correlation between known localizations and protein interactions of yeast proteins. (a) The number of interactions (inside the circles) and the fraction of interactions whose proteins share localization information (outside the circles) of three interaction databases: BiG, DIP and SGD. (b–d) The PLCPs of BiG, DIP and SGD, respectively. Given a protein at a particular localization (row), each cell corresponds to the conditional probability of the localization of its interacting partners (column). The squares on the diagonal (or off-diagonal) indicate the locations with relatively low (or high) degrees of location-sharing interactions within (or between) locations; the dotted circles on the diagonal indicate different patterns among three interaction databases for proteins in the lipid particle.