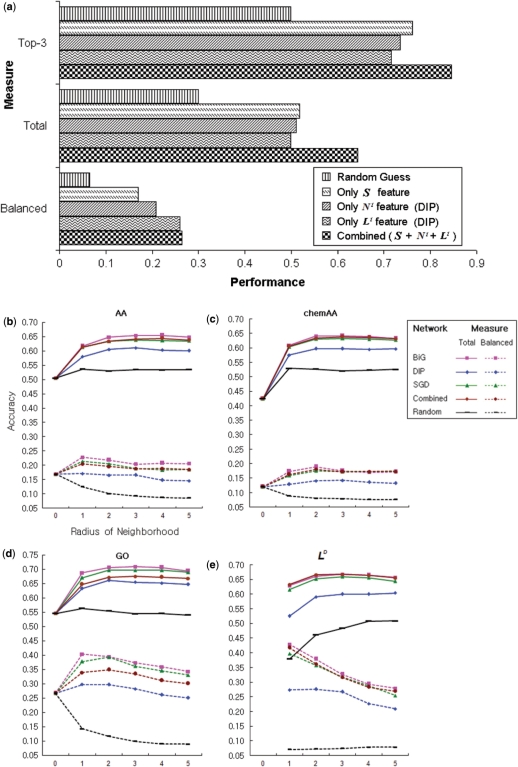
Figure 3.
Usefulness of protein interaction networks. (a) The performance of five cases, including (i) random guess of localization, (ii) S features only, (iii) N1 only, (iv) L1 only and (v) all three kinds of features. (b–e) The performance of the ND features for amino acid frequencies (b), chemical amino acid properties (c), and GO terms (d) as well as performance of the feature LD (e). Performance is based on the five interaction networks BiG, DIP, SGD, Combined, and Random (different color curves). The performance of other ND network features is shown in Supplementary Figure S3. The x-axis is the radius of neighborhood D; D = 0 means only the single protein feature vector S was used, which is a conventional approach. For Combined, the three interactome datasets BiG, DIP and SGD were pooled into a single network. For Random, localizations were randomly assigned on the BiG network. The solid lines and the dotted lines represent the Total and Balanced measures, respectively.