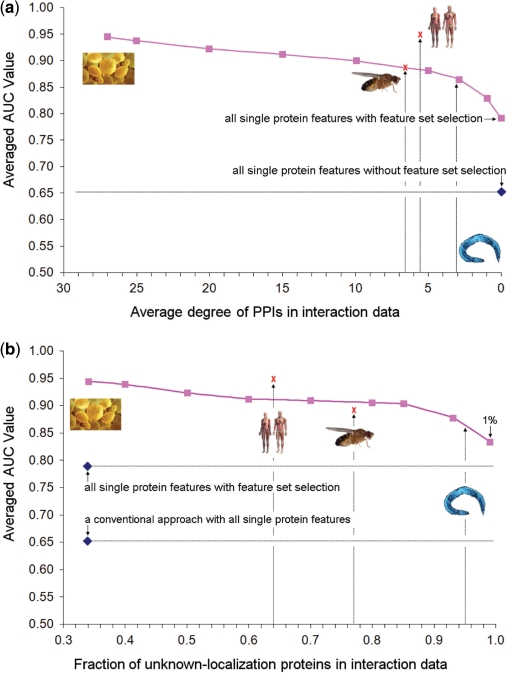
Figure 6.
Performance of predicting yeast protein localization as the available interaction (a) or localization (b) data are eroded. In (a), interactions were randomly deleted to reduce the average degree of the yeast PPI network to that specified (x-axis). In (b), known yeast protein localizations were randomly deleted. In either case, AUC was estimated using the leave-one-out approach. To avoid over fitting, the selected feature sets were taken from Supplementary Figure S6 and not re-optimized. Worm, fly, and human were mapped onto these yeast performance curves using the average degree of their available protein networks (a) or the fraction of known localizations for network proteins (b). The blue diamond represents the performance of a conventional approach using all nine single protein features without feature set selection. The red ‘X’ marks denote the performance of the proposed method when applied to recover known protein localizations in fly and human, using LOOCV.