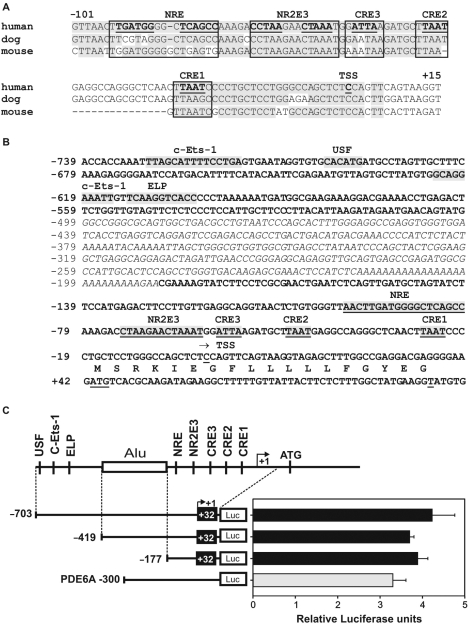
Figure 1.
Analysis of conserved transcription factor binding sites in the RS1 promoter and delineation of the proximal region. (A) Sequence alignment of the putative RS1 promoter regions from human, dog and mouse. Conserved sequences are highlighted in gray and canonical binding sites for CRX (CRE1 to 3), NR2E3 and NRL (NRE) are boxed. (B) DNA sequence of the putative human RS1 promoter. Potential cis-elements are indicated. An interspersed ALU repeat is italicized. (C) Basal activity of three RS1 promoter–luciferase constructs (−703/+32, −419/+32, −177/+32) and of a PDE6A positive control (−300) transfected into Y79 retinoblastoma cells. RS1 promoter activities are comparable to the PDE6A promoter, which is known to be active in Y79 cells. A schematic of the RS1 promoter is shown with transcription factor binding sites and the ALU repeat (box). Each transfection was repeated three times with duplicate wells analyzed. Error bars represent the standard deviation of the mean from protein normalized luciferase activities.