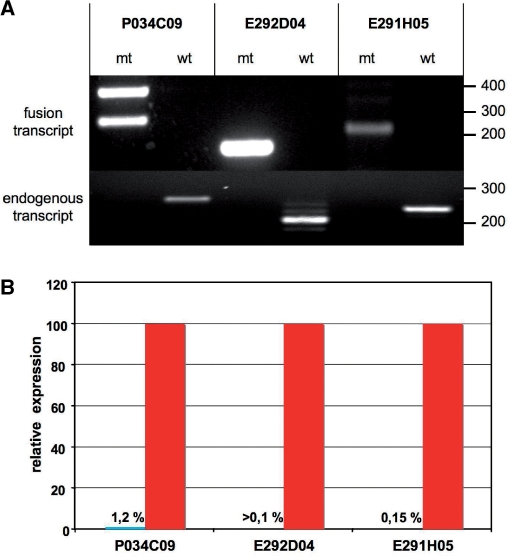
Figure 5.
Transcriptional analysis of X-linked mutations in eFlipRosaβgeo trapped ESC lines (also see Supplementary Table 1). (A) Top: RT-PCR products obtained from fusion transcripts expressed in the trapped ESC lines resolved on a 1% agarose gel stained with ethidium bromide. Amplification reactions were performed using trapped gene- and βgeo- specific primers. The double bands in lane 1 correspond to alternative N-terminal splice variants of the ATRX gene. Bottom: RT-PCR products obtained from wild-type transcripts expressed in trapped and parental E14-ESCs resolved on a 1% agarose gel stained with ethidium bromide. Gene-specific primers were chosen in exons flanking the intron containing the insertion. Note the complete absence of detectable wild-type transcripts in the trapped ESC clones. (B) Quantitative RT-PCR of the wild-type transcripts shown in A. Results are derived from triplicate reactions and were normalized to corresponding gene expression levels in E14 parental cells (=100%).