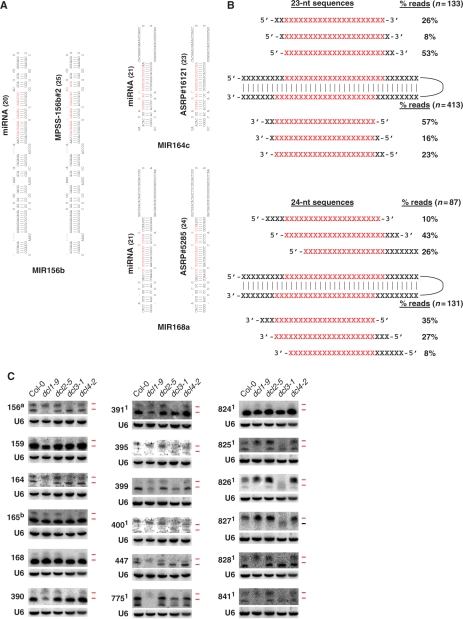
Figure 2.
Long-miRNAs represent a class of bona fide miRNAs dependent on DCL3 for their biogenesis. (A) Representative canonical miRNAs (left) and long-miRNAs (right) identified in ASRP or MPSS PLUS data sets as indicated. The miRNAs and long-miRNAs shown in red are positioned on their respective hairpin precursors. The length in nucleotides of the miRNA and long-miRNA are indicated in parentheses. The position of all remaining long-miRNAs represented in the ASRP and MPSS PLUS data sets are shown in Supplementary Figure S1A, B. (B) Cartoons showing the position of 23-nt (upper panel) and 24-nt (lower panel) long-miRNAs relative to the canonical 21-nt miRNA processed from the same hairpin. Twenty-three or twenty-four nucleotide long-miRNA sequences present in the data sets of Rajagopalan and colleagues (14) (listed in Supplementary Data S1A) were positioned with reference to the first 5′ nucleotide of either miRNA or miRNA* depending on which hairpin arm these were located. Positions (X) with identical nucleotides in the canonical and long-miRNAs are in red. The percentage of the reads with the displacement shown are indicated on the right along with the number of reads in parentheses.(C) RNA gel blot analysis of miRNAs and long-miRNAs of 18 miRNA families. SmRNAs (20 µg) from inflorescences of dcl mutants and of Col-0 wild-type were loaded on the gels. Five individual blots were probed and stripped several times. Probed miRNAs are indicated on the left side of the blots. The loading control is U6 snRNA. The upper and lower ticks to the right of each lane mark the positions of the 24- and 21-nt size markers, respectively, and are colored red when miRNA species of that size class are detected. amiR156 stands for the miR156/157 family. bmiR165 stands for the miR165/miR166. 1Indicates the miRNAs encoded by a single MIR gene.