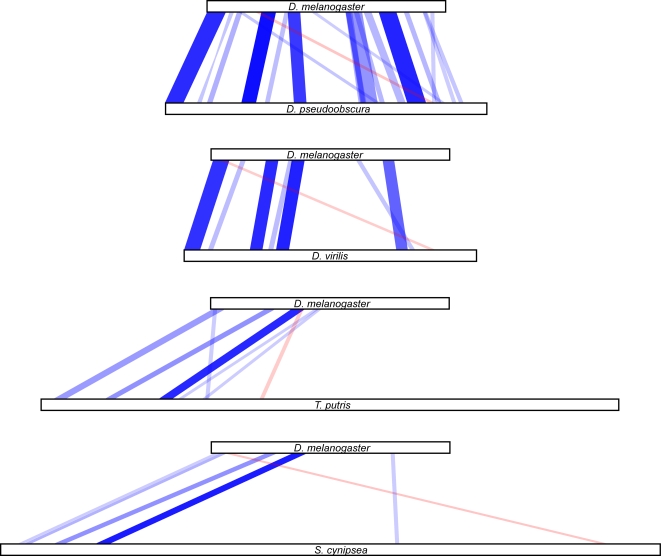
Figure 1. BLAST Similarity Maps of D. melanogaster even-skipped Stripe 2 Enhancer and Orthologous Enhancers from Drosophila and Sepsid Species.
We aligned the D. melanogaster even-skipped stripe 2 enhancer against the orthologous enhancers of D. pseudoobscura, D. virilis, T. putris, and Sepsis cynipsea (sequences as described in [1]) using NBCI BLAST bl2seq v2.2.17, with default parameters except –W (wordsize) = 9. For each species pair, high-scoring pairs (HSPs) above the default E-value cutoff of 10 were mapped by drawing a box connecting the start and end of the hit in the query and target sequence. Blue boxes represent forward strand hits, red boxes indicate reverse strand hits. The opacity of the color was scaled so that the highest scoring BLAST hits had maximal opacity of 1.0 and the lowest scoring hit had opacity of 0.1.