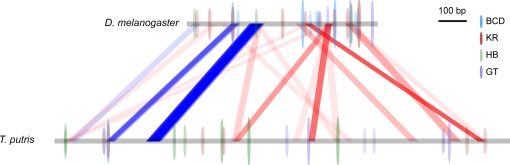
Figure 4. Similarity Map of D. melanogaster even-skipped Stripe 2 Enhancer and Predicted Binding Sites in D. melanogaster and T. putris.
We compared all 20-bp windows in the D. melanogaster and T. putris even-skipped stripe 2 enhancers (sequences as described in [1]) and mapped regions with at least 14 identical base pairs. We have found that simple percent-identity plot gives a more reliable and robust measure of similarity that BLAST. We then predicted binding sites for HB, BCD, GT, and KR in both sequences using PATSER [14] with position-weight matrixes and cutoffs for each factor as described in [1]. The height of the oval representing each predicted binding site, and the intensity of the color inside the oval, are proportional to the score of the hit.