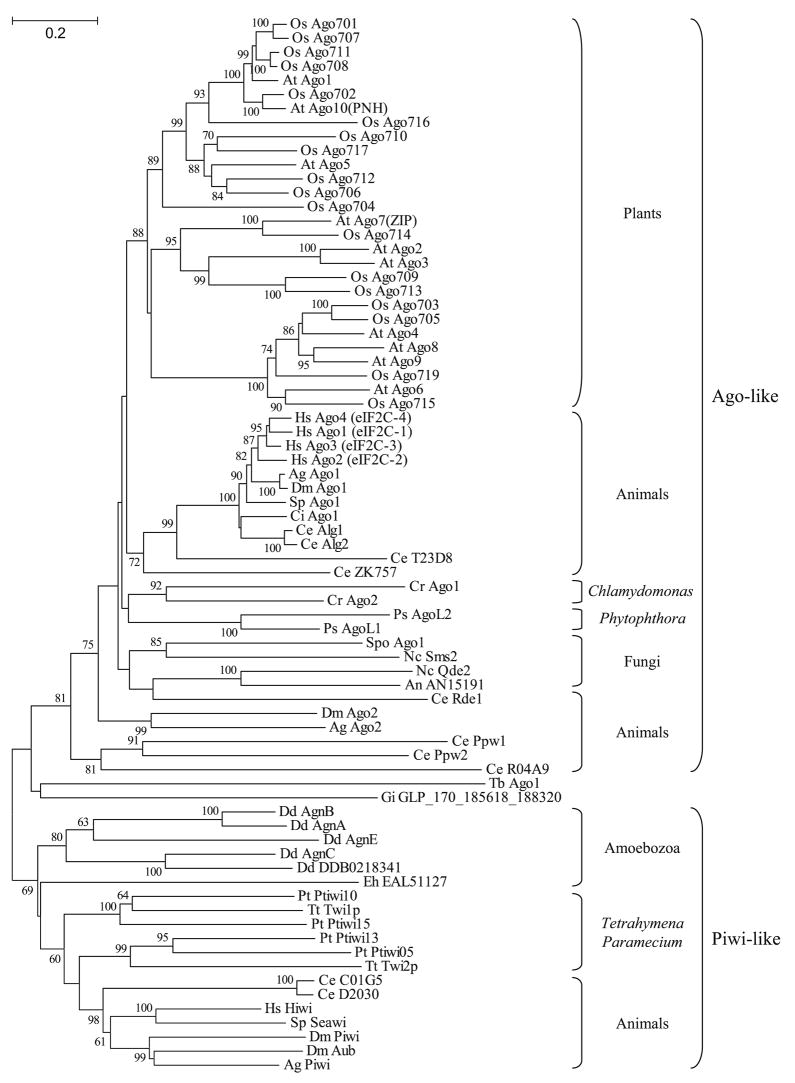
Fig. 2.
Neighbor-Joining tree showing the phylogenetic relationship among Argonaute-Piwi proteins. Sequences corresponding to the PAZ and PIWI domains were aligned using the ClustalX program and the tree was drawn using the MEGA 3.1 program. Numbers indicate bootstrap values, as percentage, based on 1,000 pseudoreplicates (only values > 60% are shown). The Ago-like and Piwi-like protein subclasses are shown to the right of the tree. Species are designated by a two-letter abbreviation preceding the name of each protein: Ag A. gambiae; At A. thaliana; An A. nidulans; Ce C. elegans; Cr C. reinhardtii; Ci C. intestinalis; Dd D. discoideum; Dm D. melanogaster; Eh E. histolytica; Gi G. intestinalis; Hs H. sapiens; Nc N. crassa; Os O. sativa; Pt P. tetraurelia; Ps P. sojae; Spo S. pombe; Sp S. purpuratus; Tt T. thermophila; and Tb T. brucei. Accession numbers of proteins used to draw the tree: Ag Ago1 EAA00062; Ag Ago2 EAL41436; Ag Piwi EAA05900; At Ago1 AAC18440; At Ago2 NP_174413; At Ago3 NP_174414; At Ago4 NP_565633; At Ago5 NP_850110; At Ago6 NP_180853; At Ago7(ZIP) NP_177103; At Ago8 NP_197602; At Ago9 NP_197613; At Ago10(PNH) CAA11429; An AN1519 EAA63775; Ce Alg1 NP_510322; Ce Alg2 NP_871992; Ce C01G5 AAB37734; Ce D2030 CAA98113; Ce Ppw1 NP_740835; Ce Ppw2 AAF60414; Ce Rde1 NP_741611; Ce R04A9 NP_508092; Ce T23D8 NP_492643; Ce ZK757 CAB54247; Cr Ago1 v2_C_130206*; Cr Ago2 v2_1200017*; Ci Ago1 C_chr_ 02q000291*; Dd AgnA EAL69296; Dd AgnB EAL62204; Dd AgnC EAL71514; Dd AgnE EAL62770; Dd DDB0218341 EAL66399; Dm Ago1 BAA88078; Dm Ago2 Q9VUQ5; Dm Aub CAA64320; Dm Piwi Q9VKM1; Eh EAL51127 EAL51127; Gi GLP_170_185618_188320 XP_779885; Hs Ago1(eIF2C-1) AAH63275; Hs Ago2(eIF2C-2) AAL76093; Hs Ago3(eIF2C-3) BAB14262; Hs Ago4(eIF2C-4) BAB13393; Hs Hiwi AAC97371; Nc Qde2 AAF43641; Nc Sms2 AAN32951; Os Ago701 XP_468547; Os Ago702 BAB96813; Os Ago703 NP_912975; Os Ago704 XP_478040; Os Ago705 AL606693; Os Ago706 NP_909924; Os Ago707 AP005750; Os Ago708 XP_473529; Os Ago709 XP_473887; Os Ago710 XP_469312; Os Ago711 AP004188; Os Ago712 XP_476934; Os Ago713 XP_473888; Os Ago714 XP_468898; Os Ago715 XP_477327; Os Ago716 XP_464271; Os Ago717 XP_469311; Os Ago719 AP000836; Pt Ptiwi05 CAI44468; Pt Ptiwi10 CAI39070; Pt Ptiwi13 CAI39067; Pt Ptiwi15 CAI39065; Ps AgoL1 v1_C_580101*; Ps AgoL2 v1_C_1000004*; Spo Ago1 O74957; Sp Ago1 XP_782278; Sp Seawi AAG42533; Tt Twi1p AAM77972; Tt Twi2p AAQ74967; Tb Ago1 AAR10810. *Accession numbers according to the annotated draft genomes of C. reinhardtii, C. intestinalis, or P. sojae at http://www.genome.jgi-psf.org/euk_cur1.html