Figure 3. The chato mutation disrupts Zfp568.
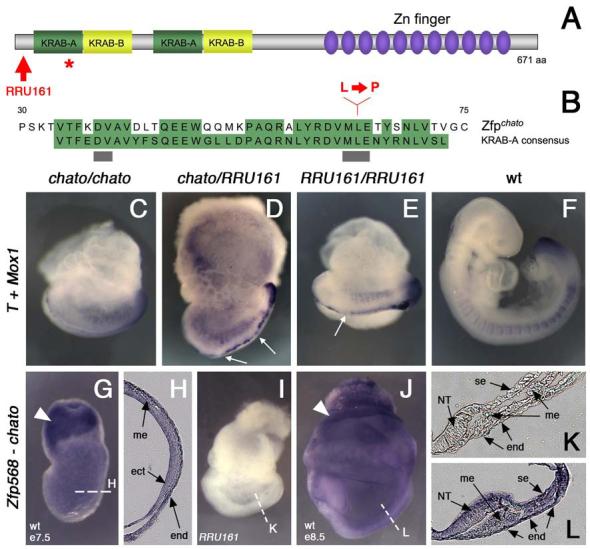
(A) Domain structure of Zfp568, which contains two KRAB-A (green)/ KRAB-B (yellow) domains and eleven Zinc fingers (purple). The red star marks the position of the chato point mutation. Red arrow points to the truncation caused by the RRU161 genetrap allele. (B) Sequence comparison of the first KRAB-A domain of Zfp568/Chato with the KRAB-A consensus sequence. Conserved residues are highlighted in green. Grey bars underline residues required for transcriptional repression (Margolin et al., 1994). Red letters indicate the Leu to Pro change caused by the chato point mutation. (C-F) Complementation test between chato and RRU161 genetrap alleles: wild type (F) and mutant embryos of the allele combinations indicated in the panels (C-E) were assayed by double in situ hybridization with T and Meox1 probes. The overall embryonic morphology, as well as defects in somites and midline, are indistinguishable between the different Zfp568 allele combinations. Notochord expression of T was irregular, showing a variable width and interruptions (arrows in D and E). (G-L) In situ hybridization with a Zfp568 probe on wild type embryos at e7.5 (G-H) and e8.5 (J, L). RRU161 mutant embryos, which generate truncated Zfp568 transcripts, were used as negative controls (I, K). Zfp568/chato is expressed in all embryonic and extraembryonic tissues, as confirmed in transverse embryonic sections (H, K-L). Zfp568 was expressed at higher levels in the extraembryonic ectoderm (solid arrowheads). (me) mesoderm, (ect) ectoderm, (end) endoderm, (NT) neural tube, (se) surface epithelia.
