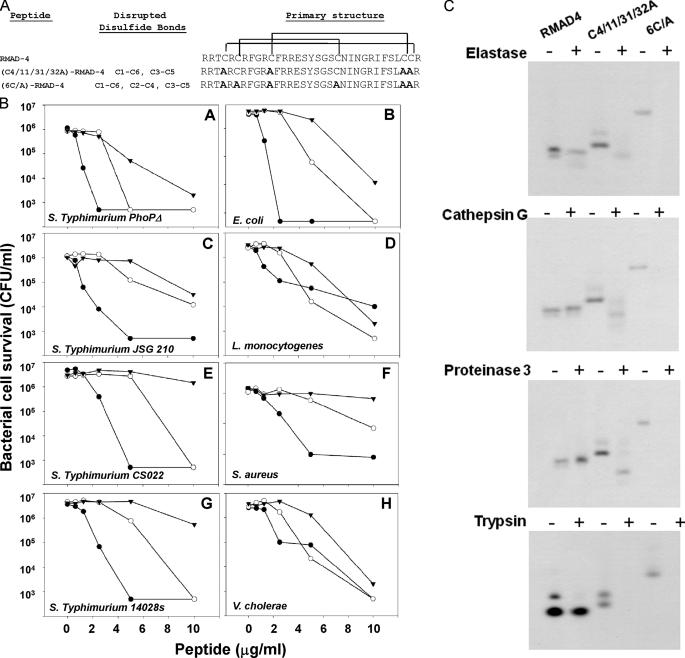
FIGURE 5.
Disulfide bonds protect RMAD-4(62–94) from degradation by serine proteases. Panel A shows the primary amino acid sequences of RMAD-4(62–94) and the two disulfide variants (C4/11/31/32/A)-RMAD-4(62–94) and (6C/A)-RMAD-4(62–94) (see “Experimental Procedures”). CI-CVI and CIII-CV disulfide bonds are disrupted in (C4/11/31/32/A)-RMAD-4(62–94) and all disulfide bonds CI-CVI, CIII-CV, and CII-CIV are disrupted in (6C/A)-RMAD-4(62–94). Mutated Cys positions converted to Ala are shown in bold font (right panel). In B, the bactericidal activities of RMAD-4(62–94) disulfide variants are compared. Exponentially growing bacterial cells were exposed to (C4/11/31/32A)-RMAD-4(62–94) (-○-), (6C/A)-RMAD-4(62–94) (-▾-), and native RMAD-4(62–94) (-•-) for 1 h at 37 °C (see “Experimental Procedures”). Both disulfide variants have activities comparable with RMAD-4 against S. typhimurium ΔphoP (A), E. coli (B), S. typhimurium JSG 210 (C), L. monocytogenes (D), and V. cholerae (H). Against S. typhimurium CS022 (E), S. aureus (F), and S. typhimurium 14028s (G), (6C/A)-RMAD-4 has attenuated activity relative to RMAD-4. C, equal quantities (10 μg) of RMAD-4, (C4/11/28/29A)-RMAD-4(62–94), and (6C/A)-RMAD-4(62–94) were digested for 18 h in the presence of NE, CG, P3, or trypsin, resolved on AU-PAGE, and stained with Coomassie Blue. Disulfide bonds protect α-defensins from proteolysis during conversion to active forms.