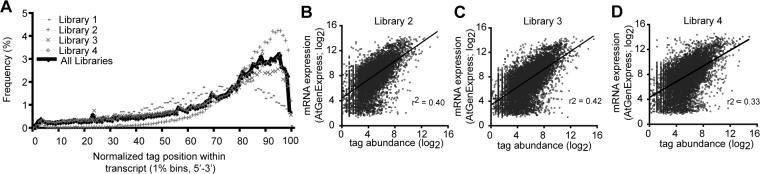
Figure 1. Degradome tags were 3' biased and correlated with transcript abundance.
(A) Histogram displaying the 5' positions of degradome tags from the indicated libraries relative to normalized transcript position. Tags were counted in one percent bins. For clarity, one tag that was extremely abundant in libraries three and four was omitted.
(B-D) Relationships between microarray-derived estimates of transcript abundance (y-axis) and degradome tag abundance (x-axis) for the indicated libraries. RMA-normalized array values were from [12] and were matched according to the tissue source (Inflorescence, ATGE_29, B and C; Seedlings, ATGE_96, D). Transcripts with degradome tag abundances equal to or less than one were omitted. Lines represent best-fit linear regressions; r2 values represent correlation coefficients.