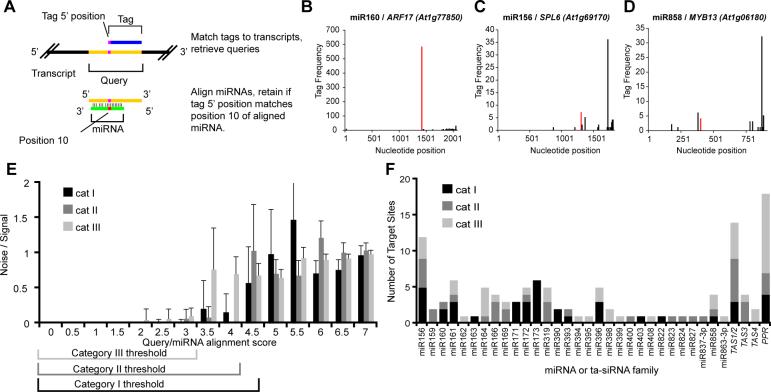
Figure 2. Experimental identification of cleaved miRNA targets without predictions.
(A) Schematic of methodology to find evidence of miRNA-mediated cleavage from degradome tags.
(B) Density of 5' position of degradome tags corresponding to ARF17, a category I target. Tags aligned with the ninth through eleventh nucleotides of a miR160 complementary site were combined and shown in red.
(C) As in B for SPL6, a category II target of miR156.
(D) As in B for MYB13, a category III target of miR858.
(E) Histogram displaying mean ratio of targets found using 30 cohorts of randomly permuted miRNAs to the number found using the annotated mature miRNA query data set at different alignment scores. Error bars represent one standard deviation. Alignment score thresholds are indicated for category I, II, and III targets.
(F) Summary of 121 cleaved miRNA and/or ta-siRNA target sites found using degradome analyses. Details in Supplementary Table 2.