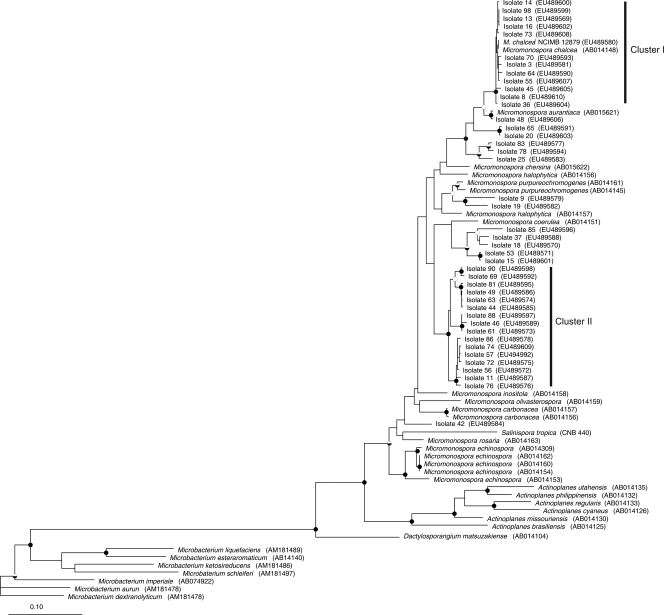
FIG. 3.
Maximum likelihood tree constructed with gyrB gene sequences from Micromonospora isolates, named strains, and other members of the Micromonosporaceae. The tree topology generated by both maximum likelihood and maximum parsimony methods was strongly supported by bootstrap values which were similar for both methods. The bootstrap values are represented by the circles in the branch nodes: filled circles, bootstrap values of >95% for both methods; half-filled circles, bootstrap values of >95% by only one method; open circles, bootstrap values of 75 to 95% for both methods. The tree was rooted with Microbacterium spp. as the outgroup; sequence accession numbers are in parentheses at the end of each branch. The scale represents 0.1 substitution per nucleotide position.