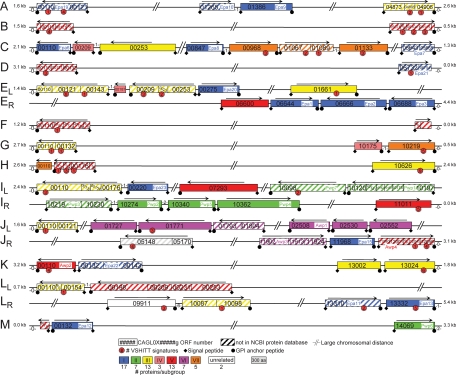
FIG. 3.
Genomic organization for adhesin-like proteins encoded in the genome of C. glabrata ATCC 2001. GPI-modified adhesin-like proteins were primarily identified by a genome-wide in silico analysis, as described previously (29). Additional adhesin-like proteins were found by pattern searching using the conserved VSHITT motif, by BLAST analysis, and by analysis of telomeric regions, where most of the adhesin-like proteins are located. Chromosomes and ORFs are numbered following Génolevures' systematic ORF numbering. Adjacent ORF fragments belonging to a single gene, as also indicated by the NCBI genome browser, are connected. Unannotated ORF fragments identified by BLASTX and containing N- or C-terminal signal peptides were connected to CAGL0B00110g, CAGL0B05115g, and Epa11. ORF sizes are to scale, but distances between ORFs are not. Colors indicate subfamilies I to VII, sharing homology in the N-terminal putative ligand-binding parts, as presented in Fig. 4. CAGL0L09911g and CAGL0J05170g (white) are unrelated outgroups in Fig. 4. Numbers of proteins in each subgroup are indicated. For CAGL0H00110g (group VII, orange) and CAGL0E00187g (group IV, pink), only C-terminal parts of the proteins were identified; their classification is therefore based on BLASTP analysis of these regions. Numbers of nonadhesive ORFs separating adhesive-like proteins and telomeres and distances of terminal adhesive-like proteins to telomeres are indicated. Arrows indicate directions of transcription.