Abstract
Kinetoplastid membrane protein 11 (KMP-11) has been identified as a flagellar protein and is conserved among kinetoplastid parasites, but its potential function remains unknown. In a recent study, we identified KMP-11 as a microtubule-bound protein localizing to the flagellum as well as the basal body in both procyclic and bloodstream forms of Trypanosoma brucei (Z. Li, J. H. Lee, F. Chu, A. L. Burlingame, A. Gunzl, and C. C. Wang, PLoS One 3:e2354, 2008). Silencing of KMP-11 by RNA interference inhibited basal body segregation and cytokinesis in both forms and resulted in multiple nuclei of various sizes, indicating a continuous, albeit somewhat defective, nuclear division while cell division was blocked. KMP-11 knockdown in the procyclic form led to severely compromised formation of the new flagellum attachment zone (FAZ) and detachment of the newly synthesized flagellum. However, a similar phenotype was not observed in the bloodstream form depleted of KMP-11. Thus, KMP-11 is a flagellar protein playing critical roles in regulating cytokinesis in both forms of the trypanosomes. Its distinct roles in regulating FAZ formation in the two forms may provide a clue to the different mechanisms of cytokinetic initiation in procyclic and bloodstream trypanosomes.
Trypanosoma brucei is an ancient parasitic protozoan that causes sleeping sickness in human and nagana in cattle in sub-Saharan Africa. Its transmission between the mammalian host and the insect vector (tsetse fly) results in different developmental forms that differ in both cellular morphology and major biological characteristics (28).
The cell division cycle of T. brucei, and presumably of all the other kinetoplastids as well, differs from that of yeasts and higher eukaryotes (29). Progression of the nuclear cycle must be coordinated with the division cycle of the mitochondrial DNA complex, the kinetoplast, for cell proliferation (54). The kinetoplast is closely associated with the flagellar basal body through a transmembrane tripartite attachment complex (33). The kinetoplast and the basal body are replicated and separated up to a finite distance before cytokinetic initiation in the insect (procyclic) form of T. brucei (9, 39). No cytokinetic initiation can occur without the appropriate segregation of basal bodies (9, 15, 36, 39, 42). In the bloodstream form, however, there is only a minimal kinetoplast-basal body segregation after their replication, and cytokinesis is tightly coupled with mitosis (29). Inhibition of mitosis in the procyclic form does not totally block cytokinesis and results in anucleate daughter cells with single kinetoplasts (zoids) (35), whereas blocking of mitosis in the bloodstream form prevents cytokinesis but allows additional rounds of organelle replication, thus producing gigantic polyploid cells with multiple kinetoplasts, basal bodies, and flagella (13, 24, 49). This apparent dissociation and association between mitosis and cytokinesis at two different developmental stages of the same organism may offer an opportunity for an in-depth analysis of the molecular mechanisms of cytokinesis without a deep entanglement with mitosis, as observed in other eukaryotes (29).
The flagellum also plays an essential role in cytokinesis in T. brucei (18). While the formation of a contractile actin-myosin II ring at the cleavage furrow has not been observed in trypanosomes during cytokinesis (12), accumulated evidence indicates that cytokinesis in trypanosomes initiates from the anterior end of the cell, and the mother and daughter cells are separated longitudinally along the two closely associated flagella (50). Silencing of flagellar proteins has been found to block cytokinesis on many occasions (7, 20, 37, 38). A knockdown of the flagellar protein FLA1 by RNA interference (RNAi) in both procyclic and bloodstream forms led to flagellum detachment and inhibition of cytokinesis without affecting mitosis, resulting in a multinucleated phenotype (20). Silencing of trypanin, a component of the flagellar dynein regulatory complex in trypanosomes, resulted in misregulation of flagellar beat and loss of directional cell motility without affecting growth of the procyclic form (16). However, a similar knockdown in the bloodstream form was lethal. Cytokinesis was inhibited, but multiple rounds of organelle multiplication continued, accumulating multiple flagella, nuclei, kinetoplasts, mitochondria, and flagellum attachment zones (FAZs) (37). Similarly, ablation of another flagellar protein, PFR2, produced a strong paralysis without a cell growth phenotype in the procyclic form (3, 4) but led to inhibited cell growth with accumulation of multiple nuclei and kinetoplasts in the bloodstream form (7). A knockdown of TAX-1, another flagellar protein, led to similar phenotypes in the bloodstream form but failed to result in any phenotype in the procyclic form (7). Apparently, many flagellar proteins are involved in regulating cytokinesis, with some of them playing a more critical role in the bloodstream than the procyclic form. The detailed mechanism behind this discrepancy remains unclear.
Much work has been done toward understanding the cell cycle control in trypanosomes, but knowledge about the molecular mechanisms coordinating mitosis and cytokinesis remains limited (29). An Aurora-like kinase homolog, TbAUK1, was found to control spindle formation, chromosome segregation, and cytokinesis in both forms of T. brucei, which may thus provide a link between mitosis and cytokinesis (24, 48). TbAUK1 and its associated proteins display a subcellular localization pattern typical of a chromosomal passenger complex during mitosis but translocalize to the anterior end of the cell after completion of mitosis, suggesting a function of the complex for cytokinetic initiation (23).
During the course of our work to identify TbAUK1-associated proteins, a flagellar protein, kinetoplastid membrane protein 11 (KMP-11) (GeneDB accession number Tb09.211.4512), was found (23). But its original coprecipitation with TbAUK1 from the trypanosome crude lysate was attributed to binding of the two proteins to the microtubules (23). KMP-11 was originally identified in Leishmania donovani as a membrane protein and a stimulator of T-lymphocyte proliferation (47). It was found in a wide variety of kinetoplastids (45), localizing to the flagellum and the flagellar pocket in Leishmania and the procyclic form of T. brucei (5, 45). It was also found in the subpellicular cytoskeleton structure in Trypanosoma cruzi (46). In Trypanosoma rangeli, however, it was identified mainly in the cytoplasm, the coat, the flagellum, and the flagellar pocket (10). We found that this protein localizes to the flagellum and the flagellar basal body in the two developmental forms of T. brucei and that its knockdown leads to a defect in basal body segregation, accumulation of multiple unequal-sized nuclei, inhibition of new FAZ formation, detachment of the new flagellum, and inhibition of cytokinesis in the procyclic form. Similar phenotypes were also observed in the bloodstream form after a KMP-11 knockdown, except that the new FAZ was synthesized and the flagellum remained attached. KMP-11 is thus a flagellar protein which plays an essential role in regulating cytokinesis in both forms of trypanosomes, though the exact regulatory mechanisms differ.
MATERIALS AND METHODS
RNAi.
The procyclic form of T. brucei strain 29-13 (53) was cultured at 26°C in Cunningham's medium supplemented with 10% fetal bovine serum (HyClone), 15 μg/ml G418, and 50 μg/ml hygromycin B. The bloodstream form of T. brucei strain 90-13 (53) was grown at 37°C with 5% CO2 supplied in HMI-9 medium containing 10% fetal bovine serum, 10% Serum Plus (JRH Biosciences, Inc.), 2.5 μg/ml G418, and 5 μg/ml hygromycin B. Cells were routinely diluted with fresh medium when their density reached 5 × 106/ml.
The full-length coding sequence of KMP-11 was cloned into the pZJM vector (52) for RNAi. Transfection of the procyclic strain 29-13 and the bloodstream strain 90-13 was performed as previously described (24). The transfectants were selected under 2.5 μg/ml phleomycin and cloned on an agarose plate (8). RNAi was induced by adding 1.0 μg/ml tetracycline, and cell growth was monitored by daily counting of cells.
Northern blotting.
Total RNA was purified from T. brucei cells with the TRIzol reagent. Northern blotting was performed as previously described (25). Briefly, total RNA was blotted onto nitrocellulose membrane in a 20× SSC (150 mM NaCl and 15 mM sodium citrate) solution. Northern hybridization was carried out overnight at 42°C in 50% formamide, 6× SSC, 0.5% sodium dodecyl sulfate (SDS), 5× Denhardt's solution with 0.1 mg/ml salmon sperm DNA. The same blot was used for different probes.
Flow cytometry analysis.
Flow cytometry analysis of propidium iodide-stained trypanosome cells was carried out as previously described (22, 24). Briefly, T. brucei cells were fixed in ethanol and incubated with DNase-free RNase (10 μg/ml) and propidium iodide (20 μg/ml) before flow cytometry analysis. The DNA content of propidium iodide-stained cells was analyzed with a fluorescence-activated cell-sorting scan (FACScan) analytical flow cytometer (BD Biosciences).
Epitope tagging of proteins expressed at the apparent endogenous level in T. brucei.
KMP-11 was cloned into the pC-3HA-Neo vector, which was constructed by replacing the PTP module in pC-PTP-Neo (41) with a triple hemagglutinin (HA) epitope. This vector is a T. brucei genome integration vector derived from pBluescript SK(+) (Stratagene, La Jolla, CA) and containing a triple HA epitope and a resistance marker cassette. The triple HA epitope is followed by the 470-bp 3′ flanking region from the TbRPA1 gene, which encodes the largest subunit of RNA polymerase I. The resistance marker cassette contains the neomycin phosphotransferase gene (the Neor gene) flanked by the intergenic regions of heat shock protein 70 genes 2 and 3 and of the β- and α-tubulin genes. The linearized construct was transfected into the procyclic strain 427 or the bloodstream strain 221. Stable transfectants were selected under 40 μg/ml G418 in strain 427 or 2.5 μg/ml G418 in strain 221. The proper gene incorporation was subsequently verified by PCR.
Western blotting.
Cells were spun down, resuspended in SDS sampling solution, boiled for 5 min at 100°C, and cleared by centrifugation. The lysate was fractionated by SDS-polyacrylamide gel electrophoresis, transferred onto polyvinylidene difluoride membranes, and immunoblotted with anti-HA monoclonal antibody (MAb) as described previously (22). The same blot was reblotted with anti-α-tubulin MAb as a loading control.
Immunofluorescence microscopy.
The following primary antibodies were used: the rat MAb YL1/2 (anti-tyrosinated α-tubulin) for the basal body (17, 40, 44) (Chemicon); the mouse MAb L8C4 (anti-PFR2) for the flagellum; the mouse MAb L3B2 for the FAZ (19, 51); rabbit polyclonal antibody against the cysteine-rich acidic transmembrane protein (CRAM), known to concentrate in the flagellar pocket as the flagellar pocket marker (21, 27); and fluorescein isothiocyanate (FITC)-conjugated anti-HA MAb (Sigma-Aldrich) for HA-tagged KMP-11. For anti-HA staining, anti-CRAM-anti-HA double staining, anti-HA and YL1/2 double staining in both forms, and L3B2 staining in the bloodstream form, cells were fixed with 4% paraformaldehyde, whereas for L8C4-YL1/2 double staining and L3B2 staining in the procyclic form, cells were fixed in cold methanol at −20°C. Immunostaining was performed as previously described (22). Briefly, the fixed cells were incubated with the primary antibodies at room temperature for 60 min, washed three times, and incubated with FITC-conjugated or Cy3-conjugated secondary antibodies (Sigma-Aldrich) for another 60 min at room temperature. After three more washes, cells were stained with 1.0 μg/ml of 4,6-diamino-2-phenylindole (DAPI), and the slides were mounted in Vectashield mount medium and examined under a fluorescence microscope.
RESULTS
KMP-11 localizes to the flagellum and its basal body in both forms of T. brucei.
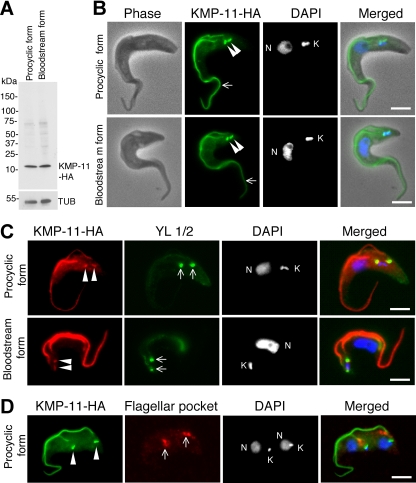
We determined the subcellular localization of KMP-11 in both procyclic and bloodstream forms of T. brucei by tagging the protein at the C terminus with a triple HA epitope and integrating the encoding DNA into one of the two alleles of KMP-11 gene. The localization of tagged KMP-11 in the transfected cells was then examined by immunofluorescence microscopy. Western blotting with anti-HA antibody indicated that the tagged protein has an expected molecular mass of ∼11 kDa in both forms (Fig. 1A). KMP-11-HA was localized to the entire flagellum throughout the cell cycle in both forms (Fig. 1B), which is consistent with observations reported in previous publications (5, 45). Additionally, discrete bright dots were also detected near the kinetoplasts (Fig. 1B to D), which overlapped well with the basal bodies stained with the YL1/2 antibody (Fig. 1C) but apparently did not overlap with the flagellar pocket (Fig. 1D), labeled by the antibody against CRAM, which is known to concentrate in the flagellar pocket (21, 27). Thus, in contrast to the previous reports, our results indicate that HA-tagged KMP-11 localizes to the flagella and the flagellar basal body, instead of the flagellar pocket.
FIG. 1.
Subcellular localization of KMP-11 in the procyclic and bloodstream forms of T. brucei. (A) Tagging of endogenous KMP-11 with the triple HA epitope in both forms. Crude cell lysates were separated by SDS-polyacrylamide gel electrophoresis, transferred onto polyvinylidene difluoride membranes, and blotted with anti-HA and anti-α-tubulin antibodies. (B) Cells expressing HA-tagged KMP-11 at the endogenous level were fixed, stained with FITC-conjugated anti-HA antibody for HA-tagged KMP-11, and counterstained with DAPI for nuclei (N) and kinetoplasts (K). Arrows indicate the flagellum staining, whereas the arrowheads point to two bright spots near the kinetoplast. (C) KMP-11 localizes to basal bodies. Cells were first immunostained with anti-HA antibody for KMP-11-HA, next stained with YL1/2 antibody for the basal body, and finally stained with DAPI. Arrowheads indicate the two bright spots of HA staining, which overlap the two YL1/2-stained basal bodies (arrows). (D) KMP-11 does not localize to the flagellar pocket. Cells were immunostained first with FITC-conjugated anti-HA antibody for KMP-11-HA and then with anti-CRAM antibody for the flagellar pocket. The two bright spots of HA immunostaining, indicated with arrowheads, do not overlie the anti-CRAM-stained flagellar pockets (arrows). Bars, 2 μm.
RNAi silencing of KMP-11 in the procyclic form inhibits cytokinesis.
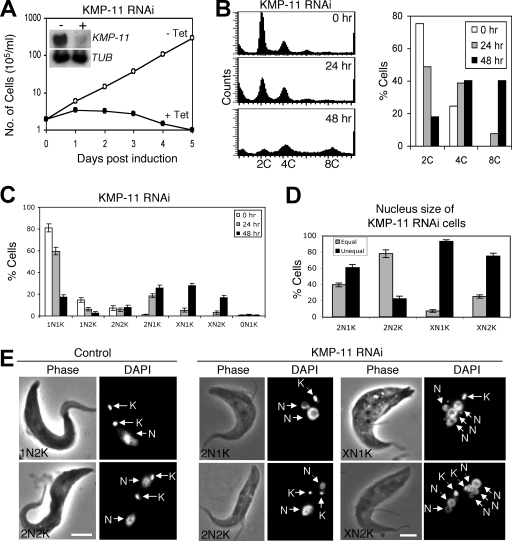
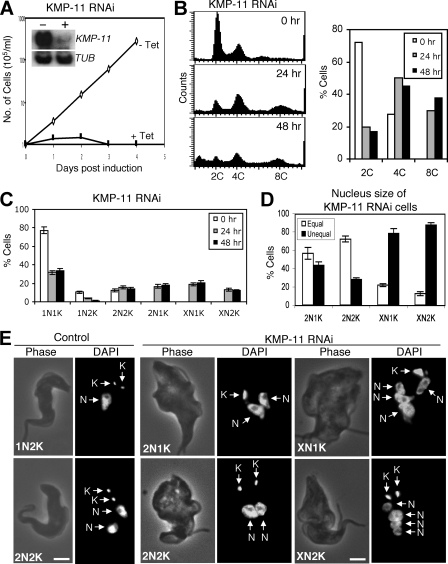
To evaluate the potential function of KMP-11, RNAi was performed in the procyclic form, and significant downregulation of KMP-11 mRNA was confirmed by Northern blotting (Fig. 2A, inset). This downregulation resulted in a dramatic inhibition of cell growth, decreased cell motility, and eventual cell death after 3 days of RNAi (Fig. 2A). Flow cytometry analysis showed that silencing of KMP-11 led to a decrease of cells with 2C DNA content from ∼75% to ∼20%, accompanied by an increase of cells with 4C DNA content from 25% to ∼40% and an accumulation of cells with 8C DNA content from 0% to ∼40% after RNAi for 48 h (Fig. 2B). These results suggest that depletion of KMP-11 inhibited cytokinesis but did not prevent additional rounds of nuclear division.
FIG. 2.
RNAi silencing of KMP-11 in the procyclic form of T. brucei. (A) A clonal cell line harboring the KMP-11 RNAi construct was cultivated with (+ Tet) or without (− Tet) tetracycline and monitored for cell growth. The inset shows the levels of mRNA, monitored by Northern blotting, in cells before (−) and after (+) tetracycline induction of RNAi for 48 h. α-Tubulin (TUB) was included as a loading control. (B) Flow cytometry analysis of DNA contents in KMP-11 RNAi cells. (C) Control and KMP-11 RNAi cells after 24 and 48 h tetracycline induction were assessed for numbers of nuclei (N) and kinetoplasts (K). Data are means ± standard errors for ∼200 cells counted in three independent experiments. (D) Percentages of 2N1K, XN1K, and XN2K cells with equal and unequal nuclear sizes after RNAi induction for 48 h. (E) Morphology of KMP-11 RNAi cells. Control and RNAi cells were stained with DAPI for nuclei (N) and kinetoplasts (K). Bars, 2 μm.
RNAi of KMP-11 in the procyclic form resulted in defective kinetoplast segregation and unequal nuclear division.
We further examined the RNAi cells by staining them with DAPI for nuclear (N) and kinetoplast (K) DNA. After a 48-h RNAi, cells with two nuclei and one kinetoplast (2N1K) increased from 0% to ∼25%, cells with multiple nuclei and one kinetoplast (XN1K) increased from 0% to ∼30%, and those with multiple nuclei and two kinetoplasts (XN2K) increased from 0% to ∼18%. This was accompanied by a decrease of 1N1K cells from ∼80% to ∼18% and 1N2K cells from ∼16% to ∼5% (Fig. 2C), suggesting that 2N1K and XN1K cells are derived from 1N1K cells and XN2K cells from 1N2K and 2N2K cells. It is thus likely that kinetoplast replication-segregation is inhibited while nuclear division continues when KMP-11 is depleted. The single kinetoplast in 2N1K and XN1K cells appears to be enlarged, indicating that kinetoplast replication may not be inhibited but its segregation is. The single enlarged kinetoplast may contain two unsegregated kinetoplasts. The two kinetoplasts in 2N2K cells are positioned closely together in the midportion of the cell body and between the two segregated nuclei, which is in striking contrast to the control 2N2K cells, in which one kinetoplast is always located at the posterior end (Fig. 2E). Similarly, the two kinetoplasts in the XN2K cells are also positioned closely together in the midportion of the cell (Fig. 2E). These results suggest that kinetoplast segregation is inhibited after knockdown of KMP-11.
Despite the fact that the nucleus continues to replicate and segregate when KMP-11 is silenced, the sizes of nuclei in 2N1K, XN1K, and XN2K cells vary significantly. After a 48-h RNAi, ∼60% of the 2N1K cells contained one large nucleus and one small nucleus, whereas only ∼20% of the 2N2K cells, which remained relatively unchanged in the population during RNAi (Fig. 2C), contained two nuclei of different sizes (Fig. 2D and E). Strikingly, ∼90% of the XN1K cells and ∼70% of the XN2K cells contained multiple nuclei of various sizes (Fig. 2D and E). Thus, only cell types with nuclei of unequal sizes underwent population increases during RNAi. These results suggest that RNAi silencing of KMP-11 also leads to unequal nuclear division.
RNAi of KMP-11 in the procyclic form inhibits basal body separation.
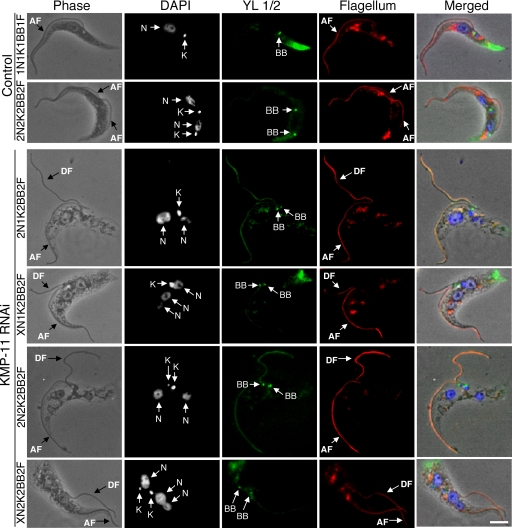
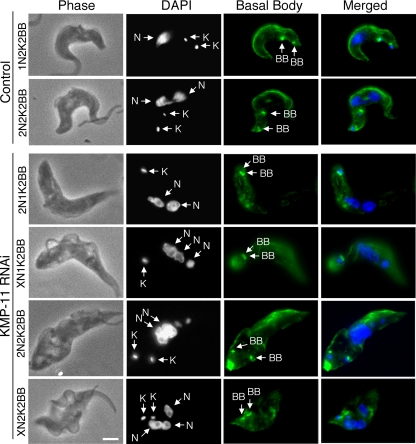
Since KMP-11 localizes to the flagellum and the basal body, we investigated the potential effect of KMP-11 RNAi on duplication and separation of the basal body as well as the growth of new flagella. The RNAi cells were fixed in cold methanol and stained with YL1/2 antibody for the basal body and L8C4 antibody for the flagellum. Both 2N1K cells and the XN1K cells were found to contain a pair of basal bodies associated with a single enlarged kinetoplast, suggesting that basal body duplication is not inhibited but segregation is blocked (Fig. 3). 2N2K and XN2K cells all contained two basal bodies, which are associated with the two kinetoplasts and positioned close together in the midportion of the cell (Fig. 3). All the cells, regardless of whether they had one or two kinetoplasts, had two closely positioned basal bodies and possessed two flagella, with one of them, most likely the newly synthesized flagellum, being detached from the cell body (Fig. 3) (see below). These results suggest that a KMP-11 knockdown leads to blocked kinetoplast segregation and defective basal body separation but does not affect the growth of a new, albeit detached, flagellum.
FIG. 3.
Effect of KMP-11 knockdown on basal body duplication and segregation and flagellum growth in the procyclic form. Control and RNAi cells after tetracycline induction for 48 h were fixed with cold methanol, stained with YL1/2 antibody for basal bodies (BB) and L8C4 antibody for flagellum, and counterstained with DAPI for nuclei (N) and kinetoplasts (K). The attached flagellum (AF) and the detached flagellum (DF) are indicated with black arrows. Bar, 2 μm. The background staining toward the posterior ends of the cells is attributed to the YL1/2 antibody, which recognizes the tyrosinated α-tubulin at the positive end of microtubules in the posterior portion of the cell.
RNAi of KMP-11 in the procyclic form inhibits formation of new FAZ.
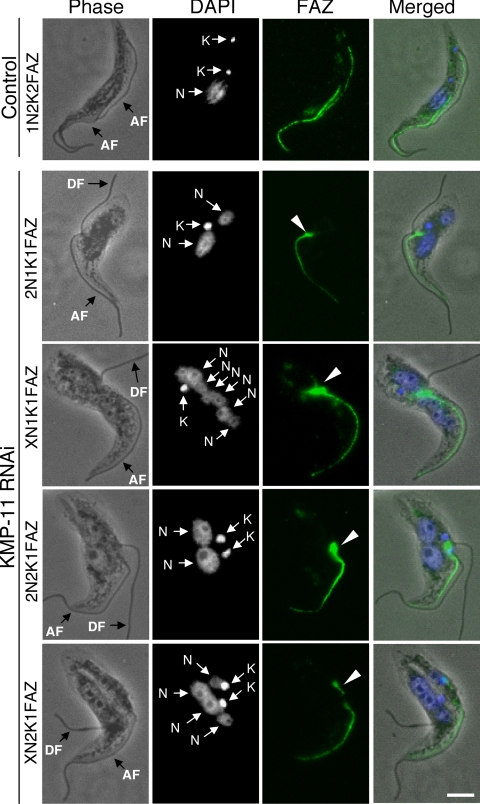
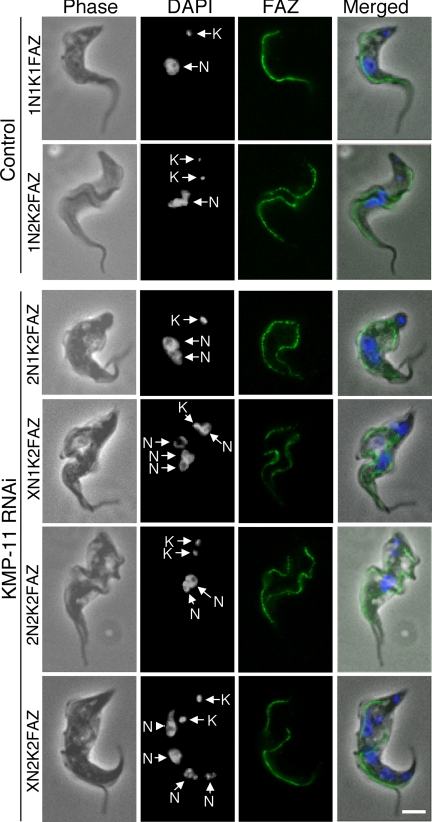
We then investigated the effect of KMP-11 knockdown on the formation of new FAZ, which is known to be essential for the attachment of newly synthesized flagellum to the cell body (51), by staining the cells with L3B2 antibody, which specifically labels the FAZ in trypanosomes. In the control 1N2K and 2N2K cells with two flagella, two FAZs of regular length were clearly detected (Fig. 4). However, for the KMP-11 RNAi cells, only a single full-length FAZ, associated with the flagellum attached to the cell body, was detected among all the 2N1K and XN1K cells and ∼90% of the 2N2K and XN2K cells (Fig. 4). These were apparently the existing FAZ and the old flagellum from the mother cell. A very short FAZ originating from the single kinetoplast in 2N1K and XN1K cells or from one of the two closely associated kinetoplasts in 2N2K and XN2K cells was, however, observed in these cells (Fig. 4). The lack of a newly formed full-length FAZ could be the cause of detachment of the newly synthesized flagellum. The mechanism behind the abortive formation of new FAZs caused by RNAi of KMP-11 is unclear.
FIG. 4.
Effect of KMP-11 knockdown on formation of new FAZs in the procyclic form. Control and RNAi cells after tetracycline induction for 48 h were fixed with cold methanol, stained with L3B2 antibody for FAZ, and counterstained with DAPI for nuclei (N) and kinetoplasts (K). The attached flagellum (AF) and the detached flagellum (DF) are indicated with black arrows. Arrowheads indicate the new, short FAZs. Bar, 2 μm.
RNAi of KMP-11 in the bloodstream form also inhibits cytokinesis.
The cell cycle control is known to differ significantly between the procyclic and bloodstream forms of T. brucei (29). We thus asked whether KMP-11 plays similar roles in the bloodstream form and the procyclic form. RNAi of KMP-11 in the bloodstream form was performed, and Northern blots showed that ∼95% of the KMP-11 mRNA was depleted after RNAi induction for 2 days (Fig. 5A, inset). This reduction of KMP-11 mRNA led to a drastic inhibition of cell growth, decrease of cell motility, and eventual cell death after 3 days of RNAi (Fig. 5A), indicating that KMP-11 is just as essential for bloodstream-form cell growth and viability as for those of the procyclic form, if not more so. We then carried out flow cytometry analysis of the control and RNAi cells and found that depletion of KMP-11 resulted in an increase in the proportion of cells with 4C DNA content from ∼30% to ∼45% and of cells with 8C DNA content from 0% to ∼40% after a 48-h RNAi (Fig. 5B), suggesting an inhibited cell division with continuous nuclear DNA synthesis, as observed in the procyclic form. Furthermore, a knockdown of KMP-11 in the bloodstream form led also to increases in proportions of 2N1K, XN1K, and XN2K cells from 0% to ∼20%, ∼23%, and ∼17%, respectively (Fig. 5C). The RNAi cells are swollen (Fig. 5E), a morphology frequently observed in the bloodstream form when cytokinesis is blocked (13, 24, 49). The single kinetoplasts in the 1K cells are enlarged, suggesting a failure in kinetoplast segregation (Fig. 5E). For the 2K cells, the sizes of kinetoplasts and their relatively short distances of separation are similar to those in the control (Fig. 5E), suggesting that these 2K cells were probably already in existence prior to RNAi induction. Sizes of the nuclei in these knockdown cells also varied significantly, as in the procyclic form (Fig. 5D). Almost 40% of the 2N1K cells, ∼80% of the XN1K cells, and ∼85% of the XN2K cells contained at least one nucleus of reduced size (Fig. 5D and E). These phenotypes indicate that KMP-11 plays similar roles in controlling cytokinesis of both bloodstream and procyclic forms of trypanosomes.
FIG. 5.
RNAi silencing of KMP-11 in the bloodstream form of T. brucei. (A) A clonal cell line harboring the KMP-11 RNAi construct was cultivated with (+ Tet) or without (− Tet) tetracycline and monitored for cell growth. The inset shows the levels of mRNA, monitored by Northern blotting, in cells before (−) and after (+) tetracycline induction of RNAi for 48 h. α-Tubulin (TUB) was included as a loading control. (B) Flow cytometry analysis of DNA contents. (C) Control and KMP-11 RNAi cells after 24 and 48 h tetracycline induction were assessed for different numbers of nuclei (N) and kinetoplasts (K). Data are means ± standard errors of ∼200 cells in three independent experiments. (D) Percentages of 2N1K, 2N2K, XN1K, and XN2K cells with equal and unequal nuclear sizes after RNAi induction for 48 h. (E) Morphology of KMP-11 RNAi cells. Control and RNAi cells were stained with DAPI for nuclei (N) and kinetoplasts (K). Bars, 2 μm.
RNAi of KMP-11 in the bloodstream form inhibits basal body separation.
Since a KMP-11 knockdown in the bloodstream form led to an apparent inhibition of kinetoplast segregation, resulting in accumulation of 2N1K and XN1K cells (Fig. 5C), we asked whether it also affects the normal basal body separation, albeit for only a relatively small distance (28). We found that in all the 2N1K and XN1K cells examined, two basal bodies were present in each cell, but they stayed very close together and associated with the single kinetoplast (Fig. 6). In 2N2K and XN2K cells, the two basal bodies were separated by a short distance, comparable to that observed in the control cells (Fig. 6). This observation supports the assumption that the kinetoplast-basal body pairs in these cells are already present and segregated prior to RNAi induction. Thus, these experiments indicate that, like in the procyclic form, a knockdown of KMP-11 in the bloodstream form blocks basal body separation.
FIG. 6.
Effect of KMP-11 knockdown on basal body duplication and segregation in the bloodstream form. Control and RNAi cells after tetracycline induction for 48 h were fixed, stained with YL1/2 antibody for basal bodies (BB), and counterstained with DAPI for nuclei (N) and kinetoplasts (K). Bar, 2 μm.
RNAi of KMP-11 in the bloodstream form has no effect on new FAZ formation and flagellum attachment.
We then looked into the possibility that a knockdown of KMP-11 in the bloodstream form may also disrupt new FAZ formation and flagellum attachment. The experimental results, presented in Fig. 7, showed that, in striking contrast to what was observed in the procyclic form (Fig. 4), two full-length FAZs were identified in all RNAi cell types and no flagellum detachment was observed in any cells examined. This is a major, and perhaps the only, distinction between the two forms of trypanosomes in terms of KMP-11 function. It suggests that KMP-11 deficiency does not prevent new flagellum synthesis in either form of trypanosomes. However, new FAZ formation is inhibited in the procyclic but not the bloodstream form. This could be useful for our further understanding of the different mechanisms of cytokinetic initiation in the two forms of trypanosomes.
FIG. 7.
Effect of KMP-11 knockdown on formation of new FAZs in the bloodstream form. Control and RNAi cells after tetracycline induction for 48 h were fixed with 4% paraformaldehyde, stained with L3B2 antibody for FAZ, and counterstained with DAPI for nuclei (N) and kinetoplasts (K). Bar, 2 μm.
DISCUSSION
In the present study, we took a new look at KMP-11 and identified its localization primarily in the basal body and flagellum in both the procyclic and bloodstream forms of T. brucei. Depletion of KMP-11 blocks cytokinesis, which could be attributed to the inhibited basal body segregation among the 1K cells of both forms. This inhibition does not affect the progression of mitosis, which continues with somewhat defective nuclear divisions. Trypanosomes do not possess typical centrioles (32). The flagellar basal bodies may constitute the microtubule-organizing centers that nucleate the flagellum (34). They are localized outside the nucleus and are not involved in spindle formation inside the nucleus during closed mitosis in trypanosomes (32). Therefore, the basal body segregation defect caused by KMP-11 knockdown (Fig. 3 and 6) likely does not affect spindle assembly directly. Indeed, several reports have indicated that inhibition of basal body duplication and/or segregation does not lead to mitotic defect in the procyclic form (15, 36, 42). In the present study, KMP-11 knockdown cells were stained with the KMX-1 antibody for the spindle structure, and this structure had no apparent alteration (data not shown). It was thus unexpected that RNAi silencing of KMP-11 would produce cells with nuclei of various sizes (Fig. 2 and 5), apparently due to defective nuclear divisions. This observation suggests that KMP-11 may play an additional role in regulating mitosis with a yet-unknown mechanism, which will make an interesting subject for further study.
The association between blocked basal body segregation and inhibited cytokinesis in the procyclic form agrees with the well-established notion that cytokinesis in the procyclic form depends primarily on the separation of basal bodies (13, 26, 35, 39). However, previous studies have also suggested that cytokinesis in the bloodstream form may not depend on basal body separation but rather is coupled primarily with nuclear division (13, 24, 49). Our current finding, showing an arrest of cytokinesis in the presence of continuing nuclear divisions in the bloodstream form, may contradict such a suggestion. It is tempting to assume that, among the 1K RNAi cells, the failure in basal body segregation, even over a relatively short distance in the bloodstream form (Fig. 6), could have caused the cytokinetic defect (Fig. 5). However, the cytokinetic inhibition in the 2K RNAi cells, in which the two basal bodies are well separated, indicates that the inhibition sets in also after basal body separation in the bloodstream form. There is thus a potential discrepancy in the role of KMP-11 in regulating cytokinesis between the two forms. While the protein may be involved in basal body segregation, it may also play a role beyond regulating basal body segregation in the bloodstream form.
Despite its localization to the entire flagellum throughout the cell cycle and its demonstrated association with the microtubules (23), KMP-11 is apparently not required for flagellum assembly, because full-length flagella are formed in both forms of trypanosomes when KMP-11 is depleted (Fig. 3 and 7). It remains to be seen, however, whether a flagellum without KMP-11 may lose some of its functions and whether such a loss may contribute to the inhibition of cytokinesis. Flagellum beating is among one of the essential features required for cytokinesis that could result from a KMP-11 knockdown (2, 6, 11). In a very preliminary examination, a loss of cell motility was noted after 2 days of KMP-11 knockdown. It remains to be seen whether this apparent motility loss could be attributed to a loss of flagellum beating.
The most intriguing finding in this report is that depletion of KMP-11 inhibited new FAZ formation in the procyclic form (Fig. 4) but not in the bloodstream form (Fig. 7) and that the absence of new FAZ may have caused the detachment of newly synthesized flagellum from the cell body of procyclic trypanosomes (Fig. 3 and 4). FAZ is a complex structure that connects the flagellum to the long axis of the cell body via the FAZ filament and the microtubule quartet with associated membrane elements (1, 43, 51). The FAZ may define the position and polarity of the cleavage plane (18) and was found to be essential for flagellum attachment and cytokinesis in the procyclic form (51). Little is known about the regulatory mechanism of FAZ synthesis. A postulated direct involvement of a basal body-flagellum protein like KMP-11 in such a function is a little difficult to comprehend at present, particularly since it functions only in the procyclic form. An easier approach to explaining the discrepant functions of KMP-11 in the two forms of trypanosomes could be to associate it with the obvious differences in triggering cytokinesis between the two forms. While the two basal bodies have to be segregated over a long distance, with one remaining in the midportion and the other migrating to the posterior end of the cell in the procyclic form, only a short-distance segregation, with both basal bodies remaining at the posterior end, is required in the bloodstream form. Since KMP-11 deficiency blocks both types of segregation, one could assume that the long-distance segregation constitutes a prerequisite for FAZ synthesis in the procyclic form, whereas the short-distance separation is not required for FAZ synthesis in the bloodstream form. Recently, RNAi silencing of three flagellum-basal body proteins (PF16, PF20, and TBBC) in the procyclic form resulted in a phenotype (1) quite similar to that displayed for KMP-11 knockdown. An analysis of the early time points in RNAi silencing of the three proteins demonstrated that the new FAZ filament, the connection of the new flagellum to the old flagellum, and base-to-tip flagellum movement in the procyclic form are all essential for basal body migration (1). This information could argue that the defect in basal body segregation caused by KMP-11 knockdown is likely due to the lack of new FAZ formation and flagellum connection in the procyclic form. However, the occurrence of a similar failure in basal body segregation in the bloodstream form, despite the presence of a full-length new FAZ, suggests a different mechanism behind the basal body segregation.
Among the functions of the other flagellar proteins that are known to be essential for cytokinesis of trypanosomes, none closely resembles that of KMP-11 except for FLA1 (20), which shares some similarities with KMP-11. FLA1 localizes mainly to the region between the cell body and the flagella (14, 31). RNAi silencing of FLA1 reduced cell growth, produced multinucleated cells, and detached flagellum in both forms of trypanosomes. But the detached flagellum forms a loop with the end of the procyclic cell, suggesting that the tip of the flagellum is still connected to the cell (30), whereas the connection is apparently lost in the KMP-11-silenced procyclic form. FLA1 is thus a protein functionally related to but not identical to KMP-11. Although FAZ formation in the FLA1 knockdown cells was not previously monitored, the generation of detached flagellar loops in these cells may suggest inhibition of FAZ formation in both bloodstream and procyclic forms as well. This constitutes an interesting difference from knocking down KMP-11 and suggests that FLA1 may be required for FAZ formation in both forms, whereas KMP-11 is needed only in the procyclic form.
Acknowledgments
We are grateful to Keith Gull of Oxford University for providing the L8C4 and L3B2 antibodies, to George A. M. Cross of Rockefeller University for T. brucei 29-13 and 90-13, to Paul T. Englund of Johns Hopkins University School of Medicine for pZJM, and to Mary G. Lee of New York University for anti-CRAM antibody. Gratitude also goes to Jack Taunton of UCSF for his kind permission to use his fluorescence microscope.
This work was supported by NIH R01 grant AI-21786.
Footnotes
Published ahead of print on 26 September 2008.
REFERENCES
- 1.Absalon, S., L. Kohl, C. Branche, T. Blisnick, G. Toutirais, F. Rusconi, J. Cosson, M. Bonhivers, D. Robinson, and P. Bastin. 2007. Basal body positioning is controlled by flagellum formation in Trypanosoma brucei. PLoS One 2e437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron, D. M., Z. P. Kabututu, and K. L. Hill. 2007. Stuck in reverse: loss of LC1 in Trypanosoma brucei disrupts outer dynein arms and leads to reverse flagellar beat and backward movement. J. Cell Sci. 1201513-1520. [DOI] [PubMed] [Google Scholar]
- 3.Bastin, P., K. Ellis, L. Kohl, and K. Gull. 2000. Flagellum ontogeny in trypanosomes studied via an inherited and regulated RNA interference system. J. Cell Sci. 1133321-3328. [DOI] [PubMed] [Google Scholar]
- 4.Bastin, P., T. Sherwin, and K. Gull. 1998. Paraflagellar rod is vital for trypanosome motility. Nature 391548. [DOI] [PubMed] [Google Scholar]
- 5.Berberich, C., G. Machado, G. Morales, G. Carrillo, A. Jimenez-Ruiz, and C. Alonso. 1998. The expression of the Leishmania infantum KMP-11 protein is developmentally regulated and stage specific. Biochim. Biophys. Acta 1442230-237. [DOI] [PubMed] [Google Scholar]
- 6.Branche, C., L. Kohl, G. Toutirais, J. Buisson, J. Cosson, and P. Bastin. 2006. Conserved and specific functions of axoneme components in trypanosome motility. J. Cell Sci. 1193443-3455. [DOI] [PubMed] [Google Scholar]
- 7.Broadhead, R., H. R. Dawe, H. Farr, S. Griffiths, S. R. Hart, N. Portman, M. K. Shaw, M. L. Ginger, S. J. Gaskell, P. G. McKean, and K. Gull. 2006. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature 440224-227. [DOI] [PubMed] [Google Scholar]
- 8.Carruthers, V. B., and G. A. Cross. 1992. High-efficiency clonal growth of bloodstream- and insect-form Trypanosoma brucei on agarose plates. Proc. Natl. Acad. Sci. USA 898818-8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das, A., M. Gale, Jr., V. Carter, and M. Parsons. 1994. The protein phosphatase inhibitor okadaic acid induces defects in cytokinesis and organellar genome segregation in Trypanosoma brucei. J. Cell Sci. 1073477-3483. [DOI] [PubMed] [Google Scholar]
- 10.Diez, H., L. Sarmiento, M. L. Caldas, M. Montilla, C. Thomas Mdel, M. C. Lopez, and C. Puerta. 2008. Cellular location of KMP-11 protein in Trypanosoma rangeli. Vector Borne Zoonotic Dis. 893-96. [DOI] [PubMed] [Google Scholar]
- 11.Gadelha, C., B. Wickstead, and K. Gull. 2007. Flagellar and ciliary beating in trypanosome motility. Cell Motil. Cytoskeleton 64629-643. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Salcedo, J. A., D. Perez-Morga, P. Gijon, V. Dilbeck, E. Pays, and D. P. Nolan. 2004. A differential role for actin during the life cycle of Trypanosoma brucei. EMBO J. 23780-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammarton, T. C., J. Clark, F. Douglas, M. Boshart, and J. C. Mottram. 2003. Stage-specific differences in cell cycle control in Trypanosoma brucei revealed by RNA interference of a mitotic cyclin. J. Biol. Chem. 27822877-22886. [DOI] [PubMed] [Google Scholar]
- 14.Haynes, P. A., D. G. Russell, and G. A. Cross. 1996. Subcellular localization of Trypanosoma cruzi glycoprotein Gp72. J. Cell Sci. 1092979-2988. [DOI] [PubMed] [Google Scholar]
- 15.He, C. Y., M. Pypaert, and G. Warren. 2005. Golgi duplication in Trypanosoma brucei requires Centrin2. Science 3101196-1198. [DOI] [PubMed] [Google Scholar]
- 16.Hutchings, N. R., J. E. Donelson, and K. L. Hill. 2002. Trypanin is a cytoskeletal linker protein and is required for cell motility in African trypanosomes. J. Cell Biol. 156867-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilmartin, J. V., B. Wright, and C. Milstein. 1982. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J. Cell Biol. 93576-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohl, L., D. Robinson, and P. Bastin. 2003. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 225336-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohl, L., T. Sherwin, and K. Gull. 1999. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J. Eukaryot. Microbiol. 46105-109. [DOI] [PubMed] [Google Scholar]
- 20.LaCount, D. J., B. Barrett, and J. E. Donelson. 2002. Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J. Biol. Chem. 27717580-17588. [DOI] [PubMed] [Google Scholar]
- 21.Lee, M. G., B. E. Bihain, D. G. Russell, R. J. Deckelbaum, and L. H. Van der Ploeg. 1990. Characterization of a cDNA encoding a cysteine-rich cell surface protein located in the flagellar pocket of the protozoan Trypanosoma brucei. Mol. Cell. Biol. 104506-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Z., S. Gourguechon, and C. C. Wang. 2007. Tousled-like kinase in a microbial eukaryote regulates spindle assembly and S-phase progression by interacting with Aurora kinase and chromatin assembly factors. J. Cell Sci. 1203883-3894. [DOI] [PubMed] [Google Scholar]
- 23.Li, Z., J. H. Lee, F. Chu, A. L. Burlingame, A. Gunzl, and C. C. Wang. 2008. Identification of a novel chromosomal passenger complex and its unique localization during cytokinesis in Trypanosoma brucei. PLoS One 3e2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Z., and C. C. Wang. 2006. Changing roles of aurora-B kinase in two life cycle stages of Trypanosoma brucei. Eukaryot. Cell 51026-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Z., and C. C. Wang. 2002. Functional characterization of the 11 non-ATPase subunit proteins in the trypanosome 19 S proteasomal regulatory complex. J. Biol. Chem. 27742686-42693. [DOI] [PubMed] [Google Scholar]
- 26.Li, Z., and C. C. Wang. 2003. A PHO80-like cyclin and a B-type cyclin control the cell cycle of the procyclic form of Trypanosoma brucei. J. Biol. Chem. 27820652-20658. [DOI] [PubMed] [Google Scholar]
- 27.Liu, J., X. Qiao, D. Du, and M. G. Lee. 2000. Receptor-mediated endocytosis in the procyclic form of Trypanosoma brucei. J. Biol. Chem. 27512032-12040. [DOI] [PubMed] [Google Scholar]
- 28.Matthews, K. R. 2005. The developmental cell biology of Trypanosoma brucei. J. Cell Sci. 118283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKean, P. G. 2003. Coordination of cell cycle and cytokinesis in Trypanosoma brucei. Curr. Opin. Microbiol. 6600-607. [DOI] [PubMed] [Google Scholar]
- 30.Moreira-Leite, F. F., T. Sherwin, L. Kohl, and K. Gull. 2001. A trypanosome structure involved in transmitting cytoplasmic information during cell division. Science 294610-612. [DOI] [PubMed] [Google Scholar]
- 31.Nozaki, T., P. A. Haynes, and G. A. Cross. 1996. Characterization of the Trypanosoma brucei homologue of a Trypanosoma cruzi flagellum-adhesion glycoprotein. Mol. Biochem. Parasitol. 82245-255. [DOI] [PubMed] [Google Scholar]
- 32.Ogbadoyi, E., K. Ersfeld, D. Robinson, T. Sherwin, and K. Gull. 2000. Architecture of the Trypanosoma brucei nucleus during interphase and mitosis. Chromosoma 108501-513. [DOI] [PubMed] [Google Scholar]
- 33.Ogbadoyi, E. O., D. R. Robinson, and K. Gull. 2003. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol. Biol. Cell 141769-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pickett-Heaps, J. 1974. The evolution of mitosis and the eukaryotic condition. Biosystems 637-48. [DOI] [PubMed] [Google Scholar]
- 35.Ploubidou, A., D. R. Robinson, R. C. Docherty, E. O. Ogbadoyi, and K. Gull. 1999. Evidence for novel cell cycle checkpoints in trypanosomes: kinetoplast segregation and cytokinesis in the absence of mitosis. J. Cell Sci. 1124641-4650. [DOI] [PubMed] [Google Scholar]
- 36.Pradel, L. C., M. Bonhivers, N. Landrein, and D. R. Robinson. 2006. NIMA-related kinase TbNRKC is involved in basal body separation in Trypanosoma brucei. J. Cell Sci. 1191852-1863. [DOI] [PubMed] [Google Scholar]
- 37.Ralston, K. S., and K. L. Hill. 2006. Trypanin, a component of the flagellar Dynein regulatory complex, is essential in bloodstream form African trypanosomes. PLoS Pathog. 2e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ralston, K. S., A. G. Lerner, D. R. Diener, and K. L. Hill. 2006. Flagellar motility contributes to cytokinesis in Trypanosoma brucei and is modulated by an evolutionarily conserved dynein regulatory system. Eukaryot. Cell 5696-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson, D. R., and K. Gull. 1991. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature 352731-733. [DOI] [PubMed] [Google Scholar]
- 40.Sasse, R., and K. Gull. 1988. Tubulin post-translational modifications and the construction of microtubular organelles in Trypanosoma brucei. J. Cell Sci. 90577-589. [DOI] [PubMed] [Google Scholar]
- 41.Schimanski, B., T. N. Nguyen, and A. Gunzl. 2005. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot. Cell 41942-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selvapandiyan, A., P. Kumar, J. C. Morris, J. L. Salisbury, C. C. Wang, and H. L. Nakhasi. 2007. Centrin1 is required for organelle segregation and cytokinesis in Trypanosoma brucei. Mol. Biol. Cell 183290-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherwin, T., and K. Gull. 1989. Visualization of detyrosination along single microtubules reveals novel mechanisms of assembly during cytoskeletal duplication in trypanosomes. Cell 57211-221. [DOI] [PubMed] [Google Scholar]
- 44.Sherwin, T., A. Schneider, R. Sasse, T. Seebeck, and K. Gull. 1987. Distinct localization and cell cycle dependence of COOH terminally tyrosinolated alpha-tubulin in the microtubules of Trypanosoma brucei brucei. J. Cell Biol. 104439-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stebeck, C. E., R. P. Beecroft, B. N. Singh, A. Jardim, R. W. Olafson, C. Tuckey, K. D. Prenevost, and T. W. Pearson. 1995. Kinetoplastid membrane protein-11 (KMP-11) is differentially expressed during the life cycle of African trypanosomes and is found in a wide variety of kinetoplastid parasites. Mol. Biochem. Parasitol. 711-13. [DOI] [PubMed] [Google Scholar]
- 46.Thomas, M. C., J. L. Garcia-Perez, C. Alonso, and M. C. Lopez. 2000. Molecular characterization of KMP11 from Trypanosoma cruzi: a cytoskeleton-associated protein regulated at the translational level. DNA Cell Biol. 1947-57. [DOI] [PubMed] [Google Scholar]
- 47.Tolson, D. L., A. Jardim, L. F. Schnur, C. Stebeck, C. Tuckey, R. P. Beecroft, H. S. Teh, R. W. Olafson, and T. W. Pearson. 1994. The kinetoplastid membrane protein 11 of Leishmania donovani and African trypanosomes is a potent stimulator of T-lymphocyte proliferation. Infect. Immun. 624893-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tu, X., P. Kumar, Z. Li, and C. C. Wang. 2006. An aurora kinase homologue is involved in regulating both mitosis and cytokinesis in Trypanosoma brucei. J. Biol. Chem. 2819677-9687. [DOI] [PubMed] [Google Scholar]
- 49.Tu, X., and C. C. Wang. 2004. The involvement of two cdc2-related kinases (CRKs) in Trypanosoma brucei cell cycle regulation and the distinctive stage-specific phenotypes caused by CRK3 depletion. J. Biol. Chem. 27920519-20528. [DOI] [PubMed] [Google Scholar]
- 50.Vaughan, S., and K. Gull. 2003. The trypanosome flagellum. J. Cell Sci. 116757-759. [DOI] [PubMed] [Google Scholar]
- 51.Vaughan, S., L. Kohl, I. Ngai, R. J. Wheeler, and K. Gull. 2008. A repetitive protein essential for the flagellum attachment zone filament structure and function in Trypanosoma brucei. Protist 159127-136. [DOI] [PubMed] [Google Scholar]
- 52.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 27540174-40179. [DOI] [PubMed] [Google Scholar]
- 53.Wirtz, E., S. Leal, C. Ochatt, and G. A. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 9989-101. [DOI] [PubMed] [Google Scholar]
- 54.Woodward, R., and K. Gull. 1990. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J. Cell Sci. 9549-57. [DOI] [PubMed] [Google Scholar]