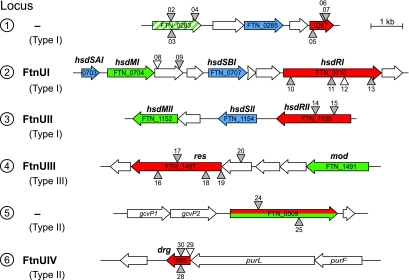
FIG. 2.
Candidate restriction loci in F. novicida. Restriction-modification genes predicted by the genome annotation (www.francisella.org) and by REBASE (rebase.neb.com) include three type I restriction genes, a type III restriction gene, a fused type II restriction and modification gene, and a type II restriction gene annotated as a dam-replacing family protein. Three additional possible restriction functions (FTN_0418, FTN_0449, and FTN_1378) are not shown but are described in the text. Putative functions are color coded: restriction (red), modification (green), specificity (blue). Gene names assigned in this work are shown in bold text above the genes. The four restriction-modification system names assigned are shown to the left. FTN_0283 (locus 1) is a modification methyltransferase pseudogene carrying a frameshift approximately two-thirds of the way through the gene (confirmed by sequencing). Numbered triangles represent transposon insertion alleles used in this study (see text and the supplemental material). Gray triangles, insertions bearing FRT site-specific recombination sites (T20 insertions); unfilled triangles, insertions without FRT sites. The orientations of the insertion within the genome are indicated by whether the triangles are shown above or below the lines representing the chromosome.