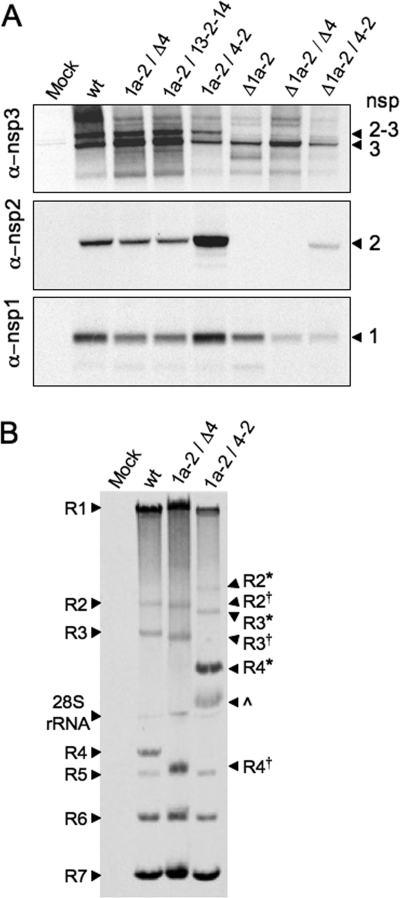
FIG. 2.
Protein expression, processing, and RNA synthesis of altered nsp2 viruses. DBT cells were infected with viruses as indicated above the gels. (A) Proteins were radiolabeled, and cell lysates were immunoprecipitated with anti-nsp1, anti-nsp2, and anti-nsp3 antibodies. “Mock” indicates mock-infected cells and “wt” indicates recombinant wild-type MHV-A59. nsps are indicated to the right of the gel: nsp2-3 (275 kDa), nsp3 (210 kDa), nsp2 (65 kDa), and nsp1 (28 kDa). (B) Viral RNA was metabolically labeled with [3H]uridine in the presence of actinomycin D from 9 to 11 h p.i. Intracellular RNA was isolated, denatured, and resolved by electrophoresis. Genomic RNA (R1) and subgenomic mRNAs (R2 to R7) are indicated. R2, R3, and R4 from the 1a-2/Δ4 virus are approximately 300 bp shorter than wild-type mRNAs and are indicated by a superscript †. The 1a-2/Δ4 virus was used as a control in which ORF4 is deleted, but its transcriptional regulatory sequence is still present, resulting in R4 comigrating with R5. R2, R3, and R4 from the 1a-2/4-2 virus, which are approximately 1,400 bp longer than wild-type R2, R3, and R4, are indicated with *. An unknown band, possibly an R4 degradation product, is indicated by a caret. R1 from wild-type and mutant viruses exhibited some variability in migration. This variability is supported by quantification of overall genomic RNA levels (1a-2/Δ4 > wt > 1a-2/4-2). RNA band sizes and quantification were determined by using ImageJ 1.40. α, anti.