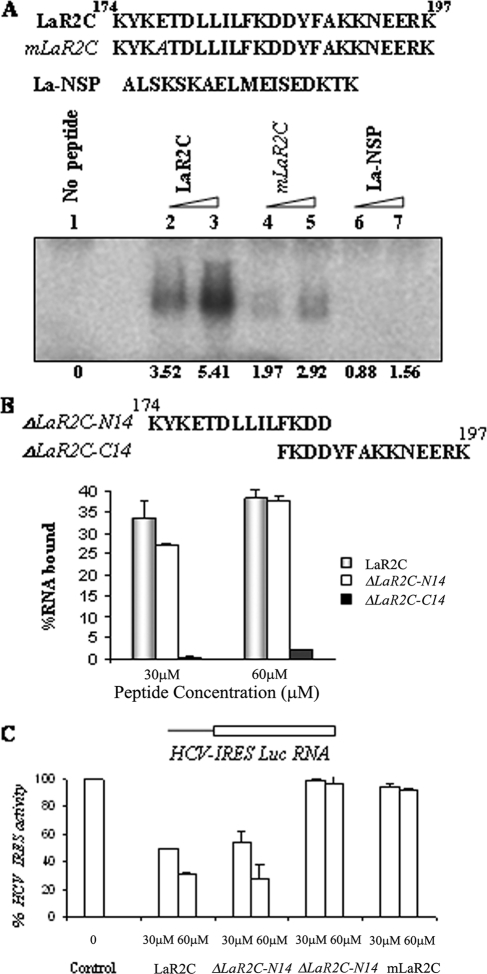
FIG. 3.
Effect of P4 point mutation in LaR2C peptide activity. (A) Schematic representation of the peptide used in the UV cross-linking analysis. The residue mutated in the mutant LaR2C peptide is indicated in italics. [α-32P]UTP-labeled HCV IRES RNA (∼75 fmol) was UV cross-linked with increasing concentrations (30 μM and 60 μM) of LaR2C, mLaR2C, and La-Nsp (NSP). The peptide-nucleotide complex was resolved in 15% Tris-Tricine PAGE followed by phosphorimaging analysis. The band intensities were quantified by densitometry. The numbers below the lanes represent the intensities, taking lane 1 (no peptide) as control. (B) Filter binding assay: 32P-labeled HCV IRES RNA was bound to increasing concentrations of either wt LaR2C peptide or mutant peptide as indicated. The amount of bound RNA was determined by its binding to the nitrocellulose filters. The percentage of bound RNA was graphically represented relative to the peptide concentration (μM). (C) Effect on HCV IRES-mediated translation in vitro. One microgram of uncapped HCV IRES Luc RNA was translated in RRL in the absence (lane 1) or in the presence of increasing concentrations (30 and 60 μM) of either wt LaR2C or mutant peptides (as indicated). The relative FLuc activities were represented as percentages of the control reaction (expressed as 100%).