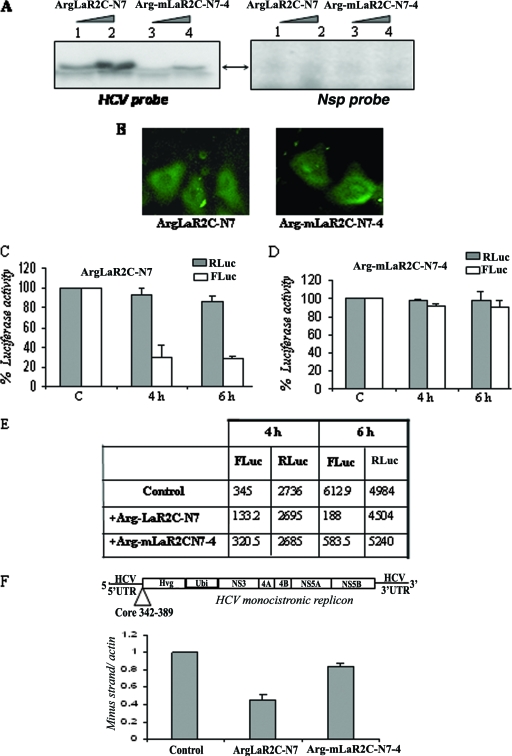
FIG. 6.
Effect of arginine-tagged LaR2C-N7 on HCV IRES function in Huh7 cells. (A) UV cross-linking. Increasing concentrations (2 μM and 4 μM) of hexa-arginine-tagged peptides, wt ArgLaR2C-N7, or the Arg-mLaR2C-N7-4 mutant were UV cross-linked with [α-32P]UTP-labeled HCV IRES RNA or a nonspecific RNA probe and analyzed by SDS-17% Tris-Tricine gel analysis, followed by phosphorimaging. (B) Fluorescein-tagged hexa-arginine peptides (both the wt and the mutant) were incubated with Huh7 cells for 3 h and washed extensively with PBS, and observed with a fluorescence microscope. Left panel shows wt ArgLaR2C-N7 and the right panel shows mutant Arg-mLaR2C-N7-4 peptide. (C and D) Huh7 monolayer cells were transfected with 2 μg of HCV bicistronic DNA (containing Rluc-HCV IRES-Fluc, in order). After 3 h of transfection, cells were overlaid with 2 μM of either wt or mutant Arg-LaR2C-N7 peptide. Cells were harvested at different time points (as indicated) and lysed, and luciferase activities were measured using the dual luciferase assay system. The RLuc (gray bar) and FLuc (white bar) activities were plotted as (fold) change increases or decreases in the presence of the peptide with respect to the corresponding control (in the absence of peptide) taken as 100. (E) Absolute values of RLuc and FLuc activities (in relative light units) of a representative experiment are presented in the table. (F) Schematic representation of the HCV monocistronic replicon RNA (9). The monolayer Huh7 cell harboring the above-mentioned replicon was overlaid with either the wt 7-mer (ArgLaR2C-N7) or mutant 7-mer (Arg-mLaR2C-N7-4) peptide (4 μM each), added twice, at 0 and 12 h. RNA was isolated at 24 h and subjected to cDNA synthesis. The HCV negative strand was detected using real-time PCR. Data were normalized with actin control, and negative strand synthesis was expressed as (fold) change compared to that of control cells (in the absence of peptide).