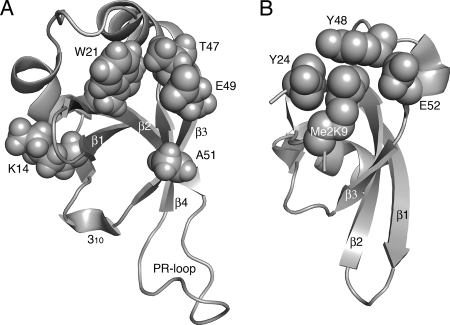
FIG. 9.
Molecular models of crucial Tudor clan member amino acid residues. (A) Amino acids critical for LEDGF/p75 PWWP domain function as defined in the present study. Residues highlighted in text are shown as space fill in the backdrop of the HDGF solution structure (31). The opening in the middle of the structure indicates the hydrophobic cavity; other structural elements are as labeled in Fig. 1B. (B) Crystal structure of the Drosophila HP1 chromodomain bound to a 15-mer histone H3 peptide dimethylated on Lys-9 (Me2K9; PBD code 1KNA). The peptide adopts β strand conformation; the sheet formed by peptide-β1-β2-β3 secondary elements (23) was aligned with β1-β2-β3-β4 from panel A. Tyr-24, Tyr-48, and Trp-45 (behind Me2K9 in this projection) form the hydrophobic binding pocket into which Me2K9 situates. A water molecule (not shown) mediates a hydrogen bond network that bridges Glu-52 backbone and side chain atoms to the dimethyl modification (23). The models were drawn by using PyMOL (10).